Meristem Genes in the Highly Reduced Endoparasitic Pilostyles boyacensis (Apodanthaceae)
- 1Departamento de Biología, Facultad de Ciencias, Universidad Nacional de Colombia, Bogotá, Colombia
- 2Instituto de Biología, Universidad de Antioquia, Medellín, Colombia
- 3Centro Nacional de Secuenciación Genómica—CNSG, Sede de Investigación Universitaria—SIU, Universidad de Antioquia, Medellín, Colombia
- 4Grupo de Parasitología, Facultad de Medicina, Universidad de Antioquia, Medellín, Colombia
- 5Instituto de Ciencias Naturales, Facultad de Ciencias, Universidad Nacional de Colombia, Bogotá, Colombia
The family Apodanthaceae comprises two genera (Apodanthes and Pilostyles) and 11 endoparasitic species, all of them lacking root and shoot apical meristems, stems, and leaves. Their vegetative phase is reduced to a mycelium-like endophyte formed by strands of parenchyma cells that are in close contact to the host vasculature. These plants become apparent only when their tiny gregarious flowers emerge breaking through the host cortex. The lack of vegetative meristems in these plants sharply contrasts to the typical formation of floral meristems. Our target species, Pilostyles boyacensis, provides a suitable system to investigate the evolution of meristem-related genes in an endoparasitic flowering plant without a typical vegetative shoot apical meristem. We have generated transcriptomes from two different developmental stages of the parasite (emerged floral buds and fruits), as well as a mixed sample that comprises the endophyte and growing floral buds of the parasite and its host, Dalea cuatrecasasii (Fabaceae). We specifically assessed copy number and domain conservation for the WUSCHEL HOMEOBOX, LRR-RLK-IX-a, and CLE families related to the maintenance of the shoot apical meristem (SAM) as well as the ARF and LFY families responsible for floral meristem identity. Five out of the 11 canonical gene families responsible for SAM maintenance and floral fate determinacy targeted in this study were found in Pilostyles. P. boyacensis shows at least one transcript with all functional domains conserved for BAM1, WUS, WOX9, ARF7, and LFY. Other genes implicated in the canonical regulatory network could not be found, including ARF5, ARF19, BAM2, BAM3, CLV1, and CLV3. In conclusion, the endoparasitic lifestyle of Apodanthaceae appears to correlate with a substantial reduction in the transcriptomic machinery linked to SAMs, whereas the genes involved in flower fate have remained intact.
Introduction
The basic body plan in vascular plants results essentially from the activity of two primary meristems that form roots [root apical meristem (RAM)] and shoots [shoot apical meristem (SAM)]. These meristems are established during early embryogenesis and are maintained during plant growth by complex genetic regulatory networks (Ha et al., 2010). In angiosperms, the SAM variously forms phytomers with photosynthetic phyllomes before its transition to flowering. Several genes are known to control the shoot meristematic activity. In Arabidopsis thaliana the WOX family gene WUSCHEL (WUS) maintains the pluripotency and cell division rate (Gaillochet and Lohmann, 2015). In turn, this protein induces the production of the short peptide CLAVATA3 (CLV3, a member of the CLAVATA 3/ESR-related genes, the CLE family), which binds to the CLAVATA1 receptor (CLV1, subfamily LEUCINE-RICH REPEAT RECEPTOR-LIKE KINASE IX-a, LRR-RLK-IX-a), triggering the negative regulation of WUS and leading to cell differentiation at the periphery of the SAM (Gaillochet and Lohmann, 2015). This finely tuned feedback loop keeps a balance between cell division and differentiation and is reinforced by the action of the receptors BARELY ANY MERISTEM 1–3 (BAM1, BAM2, BAM3; members of the LRR-RLK-IX-a). BAM receptors are expressed in the periphery of the meristematic field, where they capture other peptides of the CLE family produced on the flanks and avoid complete differentiation of the meristematic cells by the effect of CLE peptides with CLAVATA1 (DeYoung and Clark, 2008). In addition, WUSCHEL-HOMEOBOX 9 (WOX9, another member of the WOX family) has been shown to maintain active cell division in the SAM, leaf primordia initiation, and root meristem identity at early stages of seedling growth (Haecker et al., 2004; Wu et al., 2005). Thus, wox9 mutants develop an abnormal flattened SAM with differentiated cells as well as the arrest of primary root growth (Wu et al., 2005). WOX9 positively regulates WUS expression, and also responds to the negative regulation of CLV3 (Wu et al., 2005).
Hormones also play a key role in the patterning of the SAM. High concentrations of auxin on the SAM flanks are crucial for phyllome patterning, as auxin/indole-3-acetic acid (Aux/IAA) proteins interact with AUXIN RESPONSE FACTOR 5 (ARF5/MONOPTEROS, a member of the ARF family) (Korasick et al., 2014). ARF5 is repressed by IAA12/BODENLOS; such repression can be antagonized by high concentrations of auxin, triggering IAA protein degradation (Chandler, 2016). Thus, an increase of auxin in the flanks of the inflorescence meristem (IM) is necessary to release ARF5, which can directly induce LEAFY (LFY) and specify floral meristem (FM) fate (Yamaguchi et al., 2013; Denay et al., 2017). In addition, both ARF5 and ARF7 are functionally redundant in embryo axis patterning and auxin-dependent cell expansion (Hardtke et al., 2004). Furthermore, the closely related paralogs ARF7 and ARF19 could also induce LFY expression indirectly through the positive regulation of PLETHORA3 (Chandler and Werr, 2015; Taylor-Teeples et al., 2016). LFY promotes the transcription of APETALA1 (AP1) (Wagner et al., 1999) in a positive feedback loop responsible for FM identity. LFY containing complexes also activate B-, C- and E-class MADS-box genes, which ultimately control floral organ identity (Wils and Kaufmann, 2017).
Parasitic flowering plants have evolved independently at least 12 times (Nickrent, 2020). Hemiparasites are still able to photosynthesize and acquire water and minerals from its host plant. Holoparasites, instead, completely lack photosynthetic organs and relay on the host for all nutritional requirements. Such lifestyle is associated with variations of the typical body plan in response to full nutritional and developmental dependence on the host. The most dramatic reduction occurs in endoparasitic plants, which do not form roots, stems, or leaves (Heide-Jørgensen, 2008). Their vegetative phase is reduced to strands of parenchyma cells inside the host, and their only visible structures are the flowers and the fruits upon their emergence from the host tissues. Flowers frequently retain a variously modified perianth as well as a fully functional androecium and gynoecium (Heide-Jørgensen, 2008; Nikolov et al., 2013). Here, we focus on one of such extraordinary endoparasitic lineages, the Apodanthaceae. The family comprises 11 species in two genera, Apodanthes and Pilostyles (González and Pabón-Mora, 2014a, b). While the sole species of the former parasitizes members of the Salicaceae and is restricted to the New World, members of the latter grow on various legume lineages in the Americas, Africa, the Near East, and Australia (Bellot and Renner, 2014; González and Pabón-Mora, 2014a, b; Arias-Agudelo et al., 2019). Species of Pilostyles have a reduced embryo with 8–10 cells, and it is not known whether RAM or SAM formation occurs during early embryogenesis (Rutherford, 1970; González and Pabón-Mora, 2017; Figure 1M). The endophyte consists of mycelium-like cell strands in close contact with the host vasculature (Figures 1H–J). The reproductive transition occurs inside of the host passing through a bractless IM that quickly transitions into a FM. Flowers gradually break through the host cortex and emerge in preanthesis (Blarer et al., 2004; Brasil, 2010; Amaral and Ceccantini, 2011; González and Pabón-Mora, 2017; Figures 1A–G). Emerging portions are part of the exophyte (Figures 1A–F). The lack of recognizable vegetative meristems in these plants sharply contrasts to the typical formation of FMs, which poses a suitable system to study the evolution of meristem-related genes and the interplay between vegetative and FMs across flowering plants (Figures 1K,L).
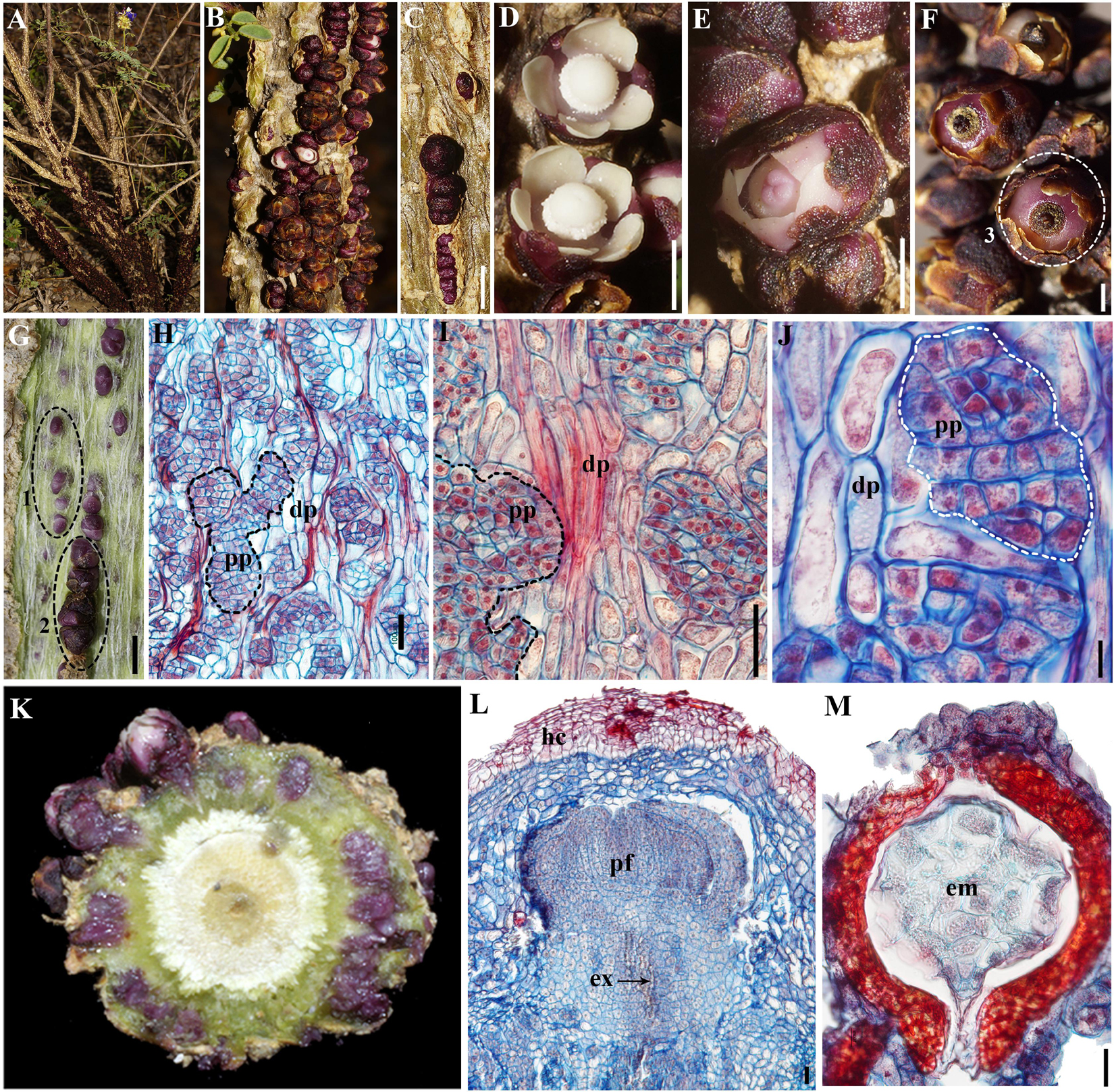
Figure 1. (A–F) Pilostyles boyacensis exophyte; (A–C) overall view; (A) detail of flowers; (B,C) flowers emerging from its host Dalea cuatrecasasii. (D) Staminate flowers; (E) carpellate flowers; (F) fruits; (G) Peeled-off stem portion of D. cuatrecasasii showing flowers of P. boyacensis at various stages of development and growth; white fibers correspond to the phloem host tissue. (H–J) Longitudinal section of infected D. cuatrecasasii stem at (H) low; (I) mid and, (J) high magnification showing irregular aggregations of Pilostyles parenchymatic cells (pp, dotted) in contact to host phloem (dp). (K) Transverse section of infected D. cuatrecasasii stem showing flowers of P. boyacensis at various stages of development and growth; (L) Longitudinal section of P. boyacensis floral bud (pf) and its vascular extensor (ex), still inside of the host cortex (hc). (M) Longitudinal section of P. boyacensis seed with embryo (em). Numbers (1–3) in (F) and (G) indicate the three tissues sampled for the transcriptomes, as follows: (1) Dalea/Pilostyles mixed tissue; (2) Preanthetic Pilostyles flowers; (3) Pilostyles fruits and seeds. Scale bars: (C–G) 2 mm; (H,I,L) 100 μm; (J) 10 μm; (M) 20 μm.
Our study generates for the first time reference transcriptomes from three different developmental time points of the endoparasitic Pilostyles boyacensis, namely, endophytic tissue of P. boyacensis growing inside the Dalea cuatrecasasii host stem, individual floral buds, and fully formed fruits of P. boyacensis. We have identified a set of genes involved in shoot apical and FMs. We provide a comprehensive list of meristem-related genes in Pilostyles and track their evolution in a broad angiosperm sampling. Altogether, the substantial reduction of the SAM genetic regulatory network affects only the formation of primary meristems, while FM development remains intact. The few SAM meristematic genes found could be functionally recruited in flower, fruit, and early embryogenesis as indicated by their expression in the P. boyacensis flowers and fruits.
Materials and Methods
Plant Material
Three samples spanning the full life cycle of the endoparasitic P. boyacensis were collected: (1) endophytic tissue of P. boyacensis growing inside the D. cuatrecasasii host stem (hereafter called PbE + D, Figure 1G); (2) individual preanthetic flowers of P. boyacensis already emerged from the host (hereafter called PbFl, Figure 1 G); and (3) young and fully formed fruits and seeds of P. boyacensis (hereafter called PbFr, Figure 1F). These samples were collected at xerophytic thickets around Villa de Leyva (Boyacá, Colombia) for total RNA extraction (Pilostyles voucher FG 4518, and D. cuatrecasasii voucher FG 4519, both deposited at COL). The tissues were flash-frozen in the field and stored in liquid nitrogen until RNA extraction was performed.
RNA Extraction, Sequencing, and Transcriptome Assembly
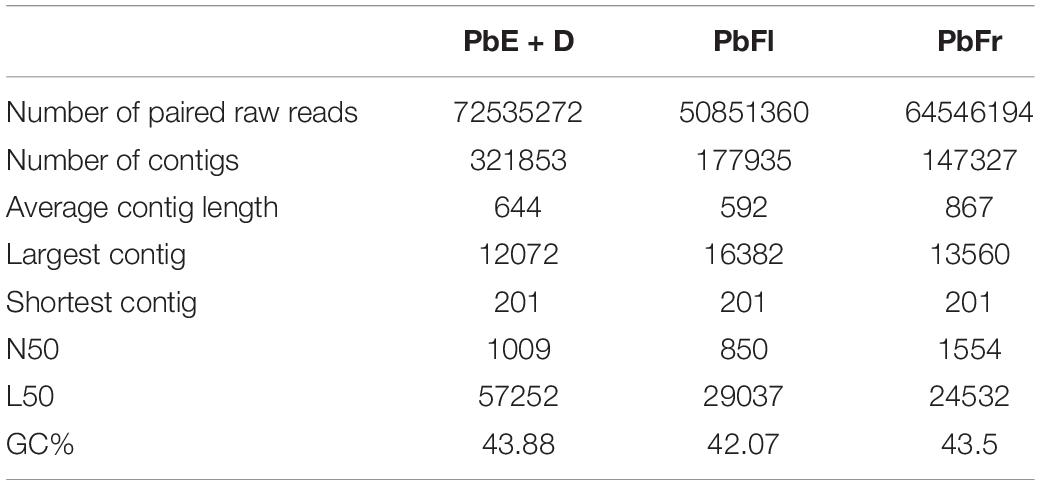
The TRIzolTM Reagent (Invitrogen) protocol was used for the extraction of total RNA with modifications used by Pabón-Mora et al. (2012). Total RNA was resuspended in 1 ml of EtOH 100% and send out to the Macrogen Sequencing Facility. The RNA-seq experiment was conducted using the Truseq mRNA library construction kit (Illumina) and sequenced on a HiSeq2000 instrument1. Read cleaning was performed with a quality threshold of Q30, and a minimum read length of 70 bp, singletons were excluded. Contig assembly was computed using the Trinity V2.5.1 software (Grabherr et al., 2011) with default settings with TRIMMOMATIC adapter removal. Assemblage metrics are summarized in Table 1.
Identification of Orthologous Genes Associated With the Regulation of Shoot Apical and Floral Meristems
The canonical genes ARF5, ARF7, ARF19, BAM1, BAM2, BAM3, CLV1, CLV3, LFY, WOX9, and WUS, involved in the SAM maintenance and FM fate in Arabidopsis thaliana, were retrieved from TAIR (The Arabidopsis Information Resource, Poole, 2007). Then, they were used as queries to search homologs in the transcriptome of Pilostyles thurberi (available in the OneKP database; Matasci et al., 2014) as well as in other members of the Cucurbitales, including the genomes of Cucumis sativus (available in Phytozome; Goodstein et al., 2012), Citrullus lanatus and Cucurbita maxima (available in Cucurbit Genomics Database; Zheng et al., 2019). These sequences were compiled and processed to keep exclusively their coding sequences (CDS). A complete matrix for each gene lineage was used as query in the transcriptomes from P. boyacensis and D. cuatrecasasii described above. BLAST searches were done using an e-value of 1 × 10–5 and 1 × 10–30. P. boyacensis genes isolated have been submitted to NCBI under the GenBank numbers MN946521–MN946537.
Sequences resulting from the search in the three Pilostyles transcriptomes were then subjected to BLASTN in NCBI. Two searches were performed in order to identify the best hits from the database and to assess their preliminary taxonomic affinity. The first search was done without any taxonomic filters. Here, all Pilostyles sequences used as queries resulted in high identity hits with genes from several taxa in the Malpighiales. The second search was done taking into consideration particular taxonomic filters, thus providing the framework to preliminary assign each sequence to either P. boyacensis or D. cuatrecasasii. This is particularly important for those sequences obtained from the mixed host-parasite transcriptome. Genes were preliminarily assigned to P. boyacensis when BLAST searches resulted in high identity and coverage values with sequences from members of Cucurbitales (where Apodanthaceae is currently circumscribed) and Malpighiales (where the first search yielded most hits). On the other hand, genes were assigned to D. cuatrecasasii if best hits matched sequences from Fabaceae (as host-related proxies). BLAST searches in Swiss-Prot (sub-base plants) were used to determine whether the sequences extracted corresponded to the target gene.
Characterization of Orthologous Genes Associated With the Regulation of Shoot Apical and Floral Meristems
In order to explore the evolution of the target genes, five independent matrices were compiled for the ARF5/7/19, CLV1/BAMs, LFY, WOX9, and WUS genes. These matrices included all sequences isolated from the three reference transcriptomes described above, as well as canonical sequences from A. thaliana, and other selected basal angiosperms, monocots, rosids, and asterids. These sequences were extracted from the GenBank, Hardwood Genomics Project2 Phytozome and OneKP. The CDSs were identified using TransDecoder3 implemented on the Galaxy platform (Afgan et al., 2018). The complete accession list is in Supplementary Table S1.
Sequences were compiled using Bioedit (Hall, 1999) and aligned in two steps: a first, unguided alignment followed by a phylogenetic analysis in order to identify major clades within large gene families such as ARF, BAM, and CLV; and a second, refined alignment guided by sequence similarity found among closely related taxonomic groups for each gene family. Alignments were made using the MAFFT server (Katoh and Standley, 2013) implementing the E-INS-I strategy, a gap opening penalty of -2 and an offset of 0.5 for most genes, except for WUS and WOX9, where a gap opening value of −3 was used. For the second alignment phase, the merge function of MAFFT was used to compile smaller datasets into a single matrix per gene family4.
In order to find the evolutionary model that best fits each matrix, ModelTest-NG (Darriba et al., 2019) was used, resulting in the GTR + G model for all gene families. Phylogenetic analyses were carried out by two approaches. First, maximum likelihood using RaxML-HPC2 (Stamatakis et al., 2008) with 500–1000 bootstrap and all other parameters by default. Second, Bayesian inference was applied using MrBayes 3.2.7a (Ronquist et al., 2012) with two independent runs with four chains, convergence between runs assessed by average standard deviation of split frequencies below 0.01, and burn-in of 25% to obtain a majority-rule consensus tree. Some features were adjusted for each gene lineage, as follows: for WUS and WOX9, three million generations were run with a sample frequency of 500; for LFY, six million generations were run with a sample frequency of 500; for CLV1/BAMs, 30 million generations were run with a sample frequency of 3,000; and for ARF5/7/19, eight million generations were run with a sample frequency of 250. The Amborella trichopoda homologs were chosen as outgroup in most phylogenetic analyses with two exceptions; the A. thaliana PHLOEM INTERCALATED WITH XYLEM (PXY) was used as outgroup in the LRR-RLK-IX-a family, given that it predates the CLV/BAM duplication (Bryan et al., 2012); and the Physcomitrella patens ARF2 was used as outgroup in the ARF5/7/19 family, as it precedes the split of ARF5 and ARF7/19 (Finet et al., 2013). Software used for the evolutionary model search, maximum likelihood and Bayesian inference were run on the CIPRES Science Gateway (Miller et al., 2010). Resulting trees were visualized and edited using FigTree v. 1.4 (Rambaut, 2006).
Unaligned aminoacidic matrices generated in Bioedit (Hall, 1999) were used to search motifs in the MEME Suite program5 with the following parameters: Motif Site Distribution: zero or one per sequence; maximum number of motifs: 50; minimum motif width: 7; maximum motif width: 150, and all other default parameters. Selected sequences including those from Pilostyles boyacensis, P. thurberi, A. thaliana, Cucurbitaceae, Salicaceae, Euphorbiaceae, and Fabaceae were subjected to MEME analysis aiming to identify domains and motifs exclusive to Apodanthaceae. A. thaliana sequences were included in order to compare the canonical domains reported for those genes. Cucurbitales sequences were included as Apodanthaceae is currently a member of this order. Finally, Salicaceae and Euphorbiaceae (Malpighiales) proteins were included due to high similarity in the BLAST hits with Pilostyles sequences. Expanded complete (in the case of WUS, WOX9, and LFY) or partial (for CLV/BAM and ARF5/7/19) datasets of the amino acid matrices were analyzed by MEME to confirm Apodanthaceae exclusive motifs.
Results
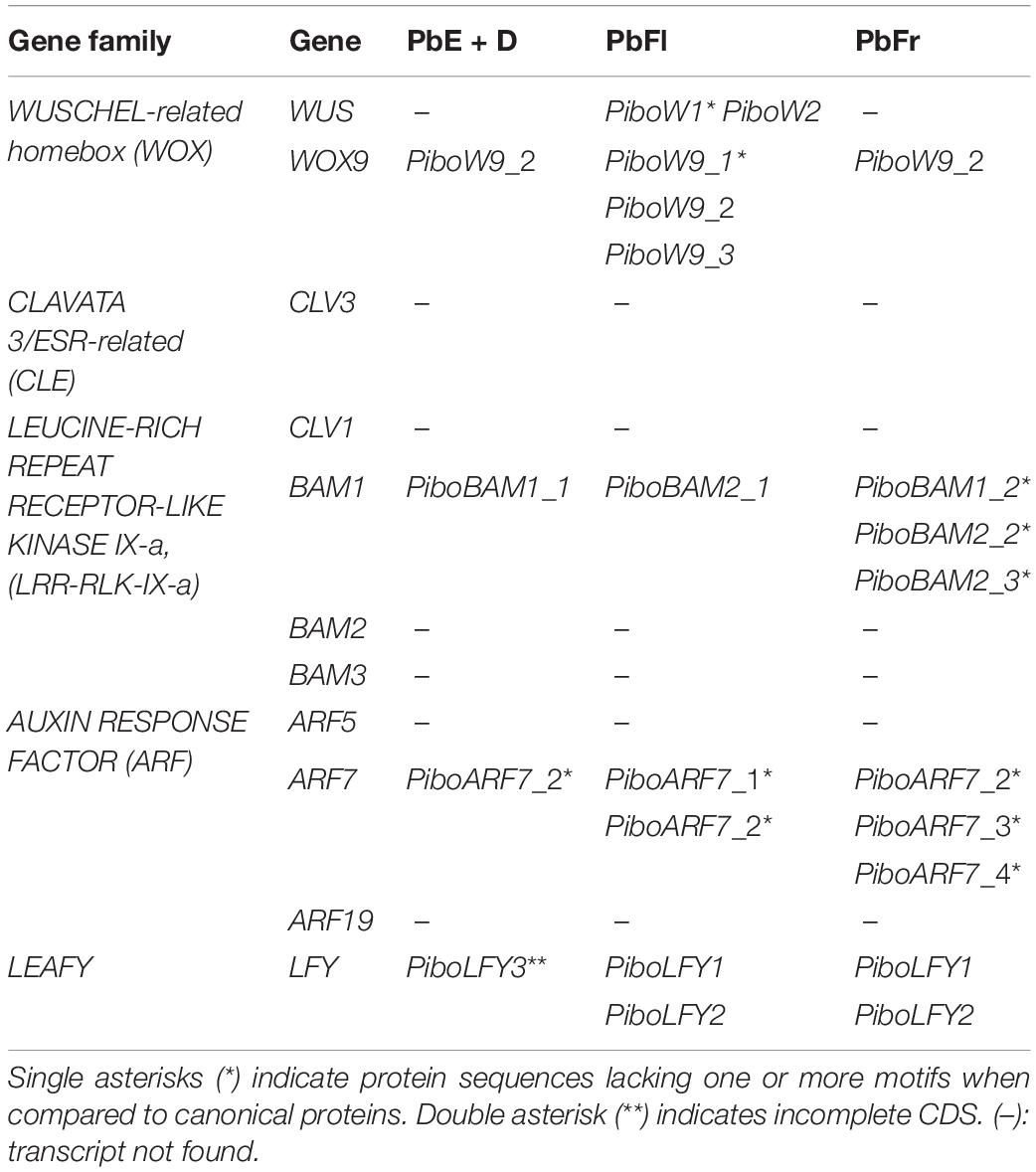
Pilostyles boyacensis orthologs of ARF7, BAM1, LFY, and WOX9 were identified from all three transcriptomes. The WUS ortholog was found only in the PbFl transcriptome. Orthologs of ARF5, ARF19, BAM2, BAM3, CLV1, CLV3 were not found in any of the three transcriptomes (Table 2). CLE22 and CLE3, both of which are CLE homologs, were only found in the PbE + D transcriptome. Their CDS size ranges between 288 and 335 nt, and they likely belong to D. cuatrecasasii, as both sequences show high identities (70–82%) and coverages (98–100%) with various Fabaceae homologs.
The WUSCHEL and WUSCHEL-Related Homeobox 9 Gene Subfamilies
The phylogenetic reconstruction of the WUS genes is mostly consistent with the systematic relationships of the angiosperm groups sampled. The only exception is the odd position of monocot homologs within rosids, likely due to long-branch attraction or convergent sites. Rosid homologs are grouped by family; thus, gene clades from Apodanthaceae, Brassicaceae, Cucurbitaceae, Euphorbiaceae, Fabaceae, Linaceae, Malvaceae, and Salicaceae are recovered (Figure 2). WUS homologs are mostly single copy genes, except for the duplicates found in Argemone mexicana (Papaveraceae), Cucurbita maxima (Cucurbitaceae), Gossypium raimondii (Malvaceae), Juglans regia (Juglandaceae), Linum usitatissimum (Linaceae), and Populus trichocarpa and Salix purpurea (Salicaceae) (Figure 2 and Supplementary Figure S1). Most duplicates appear to be species-specific, except in Salicaceae where the duplication predates the Populus/Salix diversification (Figure 2).
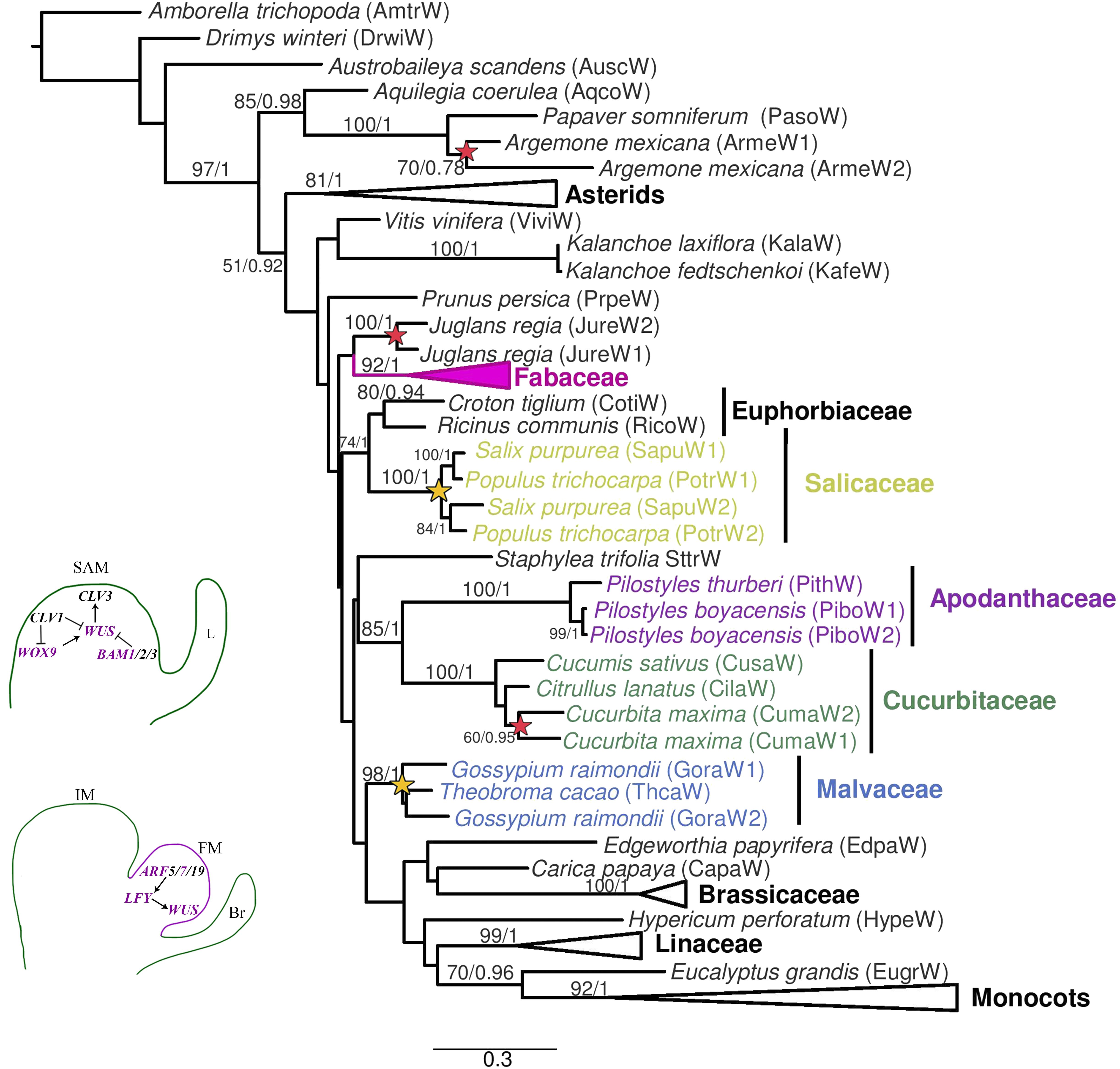
Figure 2. Maximum likelihood reconstruction of the WUSCHEL gene lineage. Bootstrap values >50 and posterior probability are shown above the nodes. Yellow stars and red stars indicate family level and species-specific duplication events, respectively. Bottom left, schematic representation of the canonical genes related to the SAM and the transition to IM and FM. Abbreviations: ARF, AUXIN RECEPTOR FACTOR; BAM, BARELY ANY MERISTEM; Br, bract; CLV, CLAVATA; L, leaf; LFY, LEAFY; WOX, WUSCHEL HOMEOBOX; WUS, WUSCHEL.
The two Pilostyles boyacensis WUSCHEL (WUS) homologs are closely related to the single sequence recovered from P. thurberi (bootstrap/posterior probability BS/PP, 100/1). WUS homologs from both Pilostyles species are, in turn, clustered with sequences from Citrullus lanatus, Cucumis sativus, and Cucurbita maxima (Cucurbitaceae) (BS/PP, 85/1, Figure 2).
The two contigs found in P. boyacensis were recovered from the PbFl transcriptome. These two differ in 10 bases and a four-base insertion that extends the reading frame by 38 amino acids toward the 3′ in one of the variants (PiboW2; Supplementary Figure S2). The MEME analysis shows that the Homeobox domain (DNA-binding domain), the LELxL WUS motif, and the TLxLFP WOX1 motif are conserved in PiboW2, while PiboW1 lacks the LELxL WUS motif (Figure 3 and Table 2). The single transcript found in the transcriptome of P. thurberi shows a premature stop codon and only conserves the Homeobox domain. Our MEME analyses did not identify additional motifs exclusive for Apodanthaceae or Cucurbitaceae.
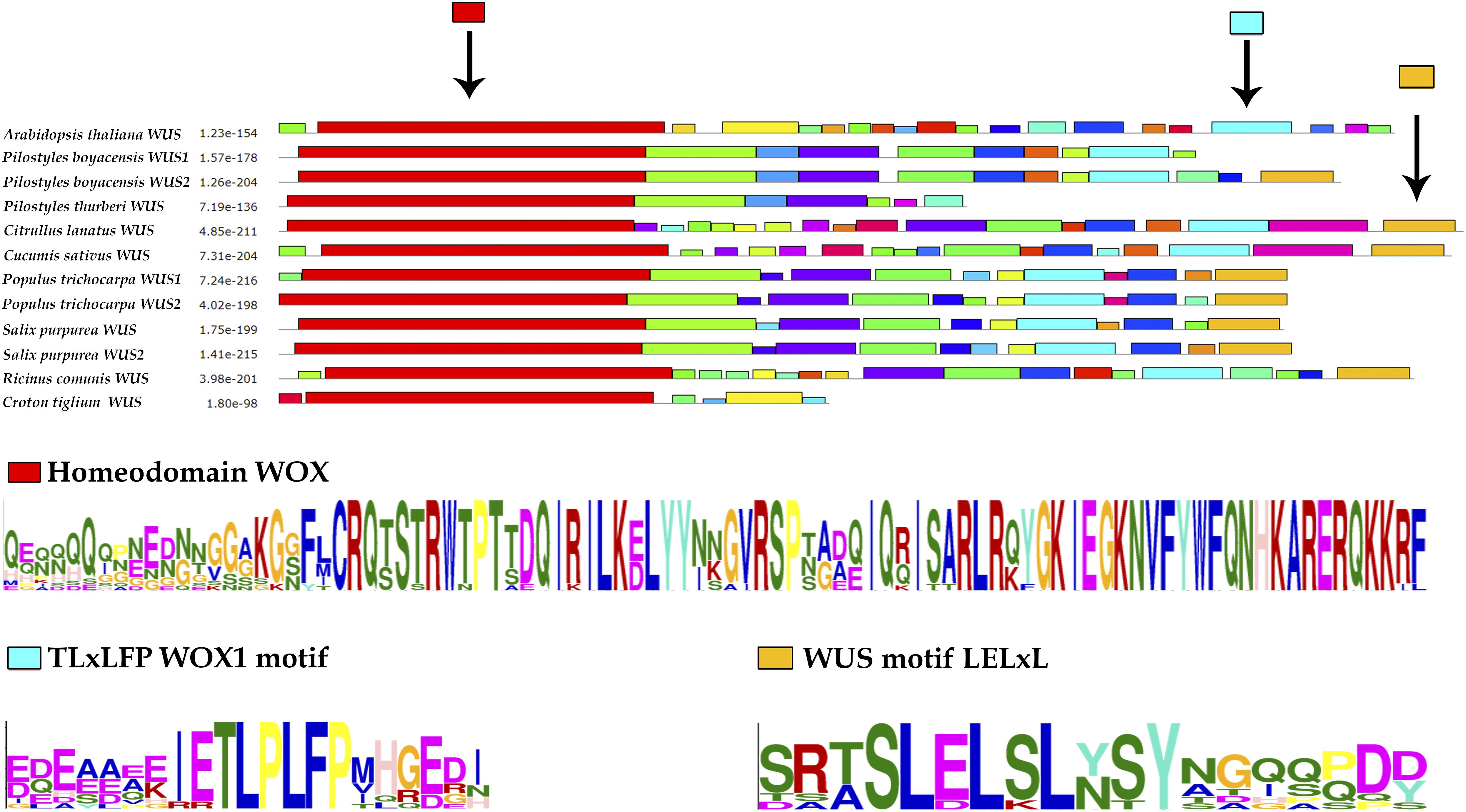
Figure 3. MEME analysis of WUSCHEL proteins of A. thaliana, Apodanthaceae, Cucurbitaceae, Salicaceae, and Euphorbiaceae.
The phylogeny of the WUSCHEL-related homeobox 9 genes recovers the monocot and the eudicot homologs with high supports (BS/PP, 86/0.98, and 70/1, respectively). However, the expected relationships within eudicot homologs are not recovered (Figure 4). Family-specific duplications were detected in Brassicaceae, resulting in the WOX8 and WOX9 clades, as well as in Crassulaceae, Poaceae and Salicaceae (Figure 4 and Supplementary Figure S3). Species-specific duplications were detected in Cucurbita maxima, Gossypium raimondii, Juglans regia, Linum usitatissimum, Musa acuminata, Pilostyles boyacensis, P. thurberi, Phaseolus vulgaris, and Staphylea trifolia (Figure 4 and Supplementary Figure S3).
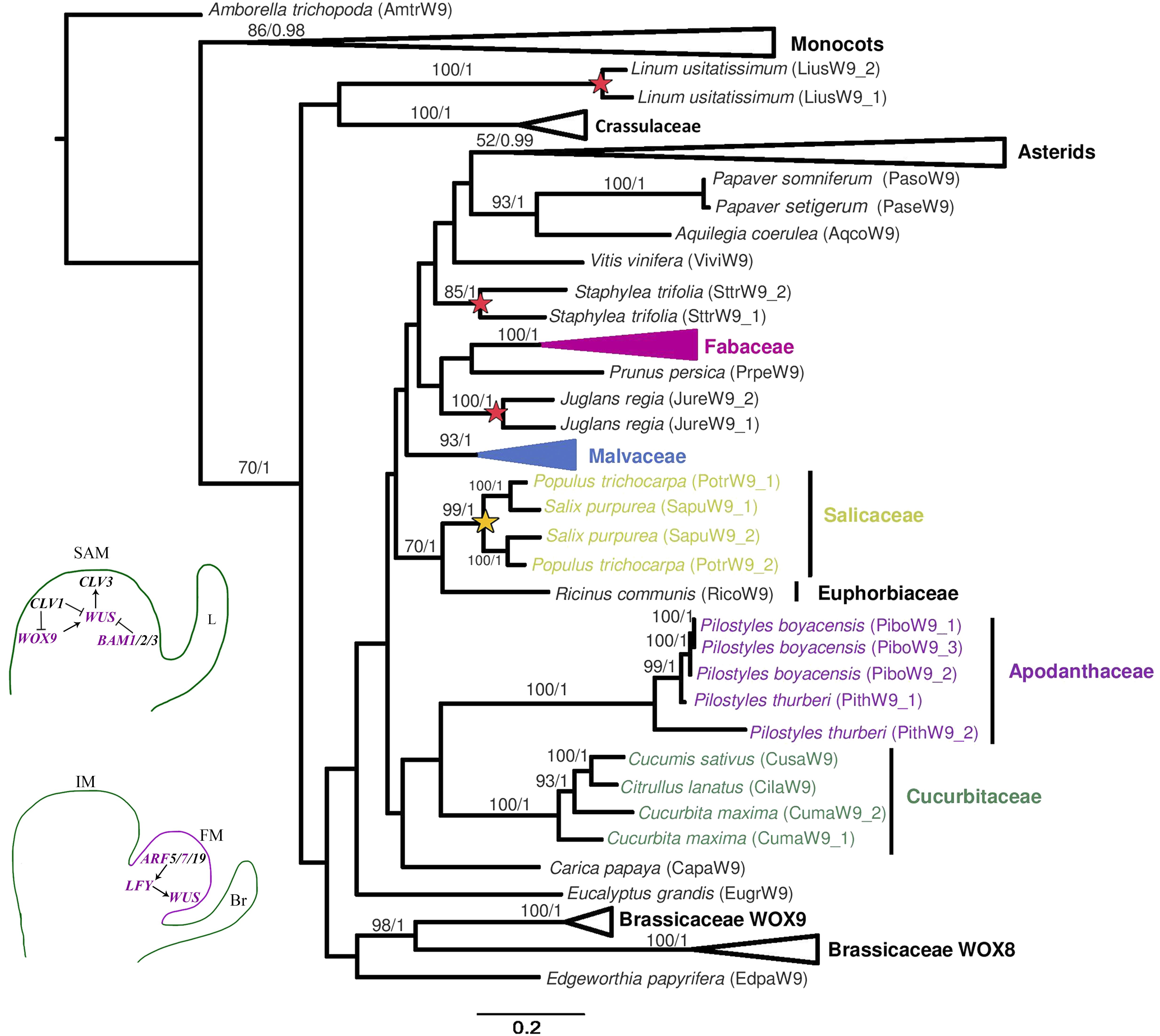
Figure 4. Maximum likelihood reconstruction of the WUSCHEL-HOMEOBOX 9 gene lineage. Bootstrap values >50 and posterior probability are shown above the nodes. Yellow stars and red stars indicate family-level and species-specific duplication events, respectively. Schemes in the bottom left follow the conventions in Figure 2.
All Pilostyles homologs cluster with Cucurbitaceae WOX 9 genes in both phylogenetic reconstructions (BS/PP, 39/0.99, Supplementary Figure S3). In the case of P. boyacensis, three variants of PiboWOX9 were detected, all of which were close to one of the two copies found in P. thurberi (PithW9_1, BS/PP, 100/1, Figure 4). The PiboW9_2 variant was found in all three transcriptomes, whereas the other two variants where only isolated from the PbFl transcriptome of P. boyacensis. The MEME analysis showed an N-terminal motif MASSN, the Homeobox domain, and the C-terminal VFIN/LQxG WOX8 motif, present in all sequences. The only exception was detected in the PiboW9_1 variant, which lacks the C-terminal VFIN/LQxG WOX8 motif (Supplementary Figure S4; Table 2). Two Apodanthaceae-specific motifs and one Cucurbitaceae-specific motif (Supplementary Figure S4) were found in a dataset that included 88 representative sequences from A. thaliana, Apodanthaceae, Cucurbitaceae, Salicaceae, and Euphorbiaceae (data not shown).
The Leucine-Rich Repeat Receptor-Like Kinase IX-a Subfamily
The phylogenetic reconstruction of the LRR-RLK-IX-a subfamily recovered four large clades CLV1 (BS/PP, 84/0.67), BAM1/2 (BS/PP, 16/-), BAM3 (BS/PP, 88/1), and BAM3-like (BS/PP, 65/1) (Figure 5 and Supplementary Figure S5). Amborella trichopoda and species of Ranunculales have homologs in each of the four clades, suggesting that duplications occurred prior to the diversification of all angiosperms. However, in 49 out of 74 species, we only detected genes from two or three clades, pointing to heterogeneous expression of the four genes in different taxa. The maximum likelihood (ML) reconstruction of CLAVATA1 subclade is mostly consistent with the phylogenetic relationships of the angiosperm groups sampled, as the A. trichopoda CLV1 homolog clusters with the sampled genes from basal eudicots, and asterid homologs separate from rosid genes (Supplementary Figure S5). The CLV1 representatives from Brassicaceae, Cucurbitaceae, Euphorbiaceae, Fabaceae, Linaceae, Malvaceae, and Salicaceae form well-supported clades (Supplementary Figure S5). Local duplications of CLV1 were detected in the host Dalea cuatrecasasii, as well as in Gossypium raimondii, Kalanchoe laxiflora, Linum usitatissimum, Prunus persica, and Salicaceae.
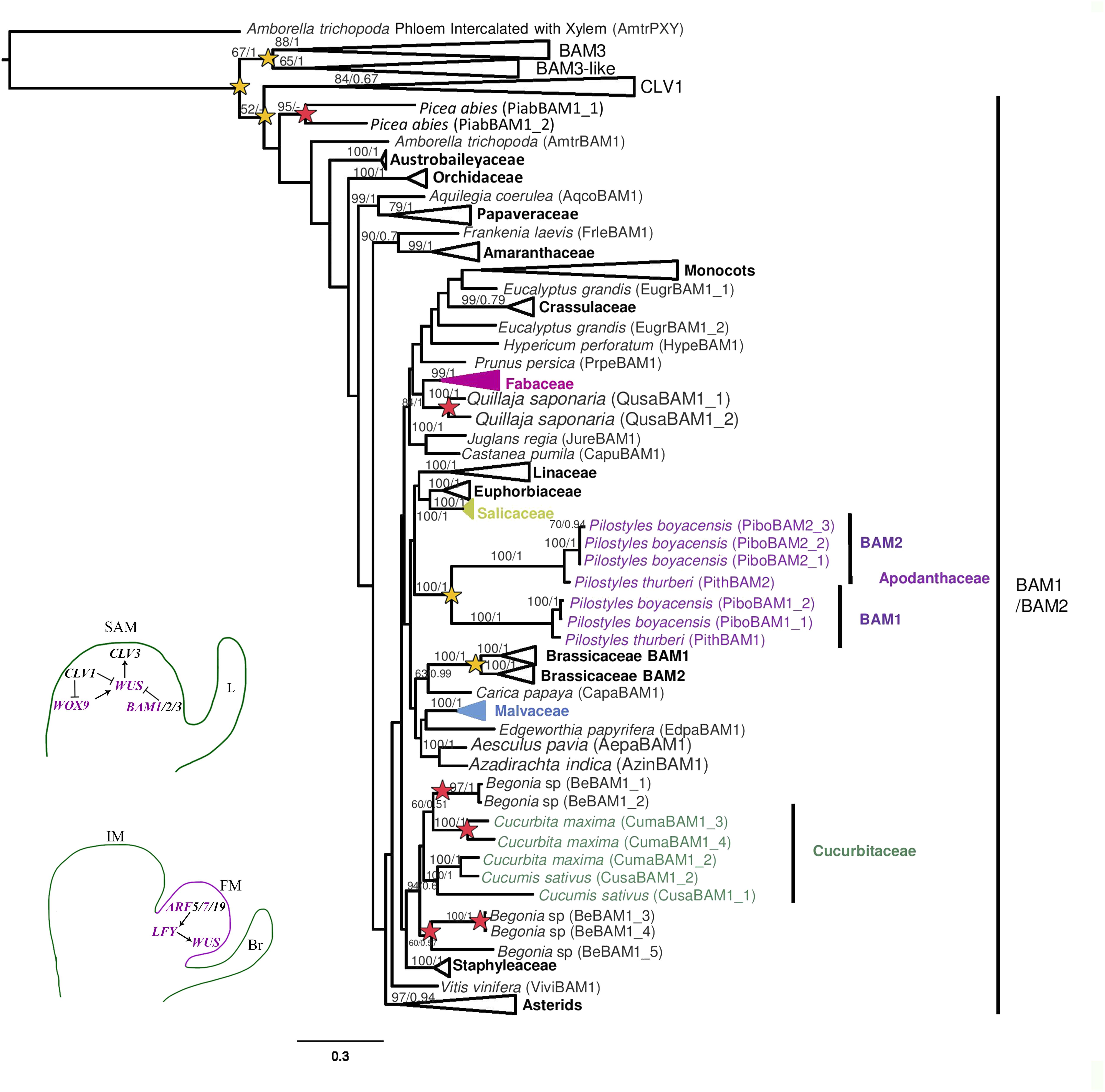
Figure 5. Maximum likelihood reconstruction of BARELY ANY MERISTEM 1-3 and CLAVATA1 gene lineages. Superclades without sequences from Pilostyles boyacensis are collapsed. Bootstrap values >50 and posterior probability are shown above the nodes. Yellow stars indicate large scale or family-level duplication events; red stars indicate species-specific duplication events. Schemes in the bottom left follow the conventions in Figure 2.
The clade formed by BAM3 and BAM3-like was retrieved in both analyses (BS/PP, 67/1). The BAM3-like subclade includes sequences from Amborella trichopoda, representative monocots (Musa acuminata) and basal eudicots (Ranunculales; Supplementary Figure S5). Asterid and rosid homologs form two separate clades; within the latter, we recovered five variants from the PbE + D transcriptome assigned using BLAST to Dalea cuatrecasasii (Supplementary Figure S5). The BAM3 phylogenetic reconstruction mirrors the taxonomic relationships among the angiosperm groups sampled (Supplementary Figure S5). The BAM3 subclade recovers family-level duplications in Fabaceae and Salicaceae, as well as species-specific duplications in Atropa belladonna, Brassica rapa, D. cuatrecasasii, and Linum usitatissimum (Supplementary Figure S5).
The ML reconstruction of BAM1/2 genes includes sampling from gymnosperms and angiosperms, and like in previous analyses, it is consistent to the phylogenetic relationships of the taxa sampled, with the exception of the position of the monocot genes close to Eucalyptus grandis (Myrtaceae) sequences (Figure 5). Within eudicots, most asterid genes cluster together (BS/PP, 97/0.94). Family-level duplications are recovered in Apodanthaceae (BAM1/2), Brassicaceae (BAM1/2), Linaceae, Papaveraceae, and Poaceae. Species-specific duplications were detected in Amaranthus hypochondriacus, Begonia sp., Chenopodium quinoa, Cladrastis lutea, Cucurbita maxima, Digitalis purpurea, Gossypium raimondii, Linum usitatissimum, L. perenne, Lonicera japonica, Medicago truncatula, Orobanche fasciculata, and Staphylea trifolia (Supplementary Figure S5). Species from Salicaceae and Euphorbiaceae show only one copy of BAM1/2 (Supplementary Figure S5). Apodanthaceae homologs are sisters to gene representatives from Salicaceae, Euphorbiaceae, and Linaceae, but support values are low (BS/PP, 35/0.67, Supplementary Figure S5). BAM1/2 homologs from P. boyacensis clustered together with those from P. thurberi (BS/PP, 100/1, Figure 5).
The MEME analysis for selected sequences within the BAM1/2 gene clade detected four leucine-rich regions, two of which were identified using Pfam as LRR4 and LRR8 (both corresponding to the extracellular domain). Additionally, two motifs matched the kinase domains (corresponding to the cytoplasmic domain). The P. boyacensis PiboBAM1_2, PiboBAM2_2, PiboBAM2_3 from PbFr, and the C. sativus CusaBAM1.1 variants lack half of the protein kinase domain (Supplementary Figure S6; Table 2). Nevertheless, PiboBAM1_1 isolated from PbE + D, and PiboBAM2_1identified in the PbFl transcriptome, as well as CusaBAM1.2, exhibit complete domains (Supplementary Figure S6; Table 2). No motifs exclusive for Apodanthaceae, Cucurbitaceae, or Cucurbitales were found.
The Auxin Response Factor 7 Gene Family
The phylogenetic reconstruction of the ARF5/7/19 gene lineages recovers five clades corresponding to ARF7-A (BS/PP, 99/1), ARF7-B (BS/PP, 61/0.99), ARF5 (BS/PP, 68/0.88), ARF7/19-D (BS/PP, 97/1), and ARF7-E (BS/PP, 92/1, Figure 6). The Amborella trichopoda and the Austrobaileya scandens homologs are found in four and three of the five clades, respectively (Supplementary Figure S7). This suggests that duplications occurred before the radiation of all angiosperms. Both ARF7-A and ARF7-B gene phylogenies are consistent with the phylogenetic relationships of the taxa sampled. Within ARF7-A no family-level duplications are detected. Only species-specific duplications are found in Argemone mexicana, Linum perenne, Orobanche fasciculata, Papaver somniferum, and Papaver setigerum (Supplementary Figure S7).
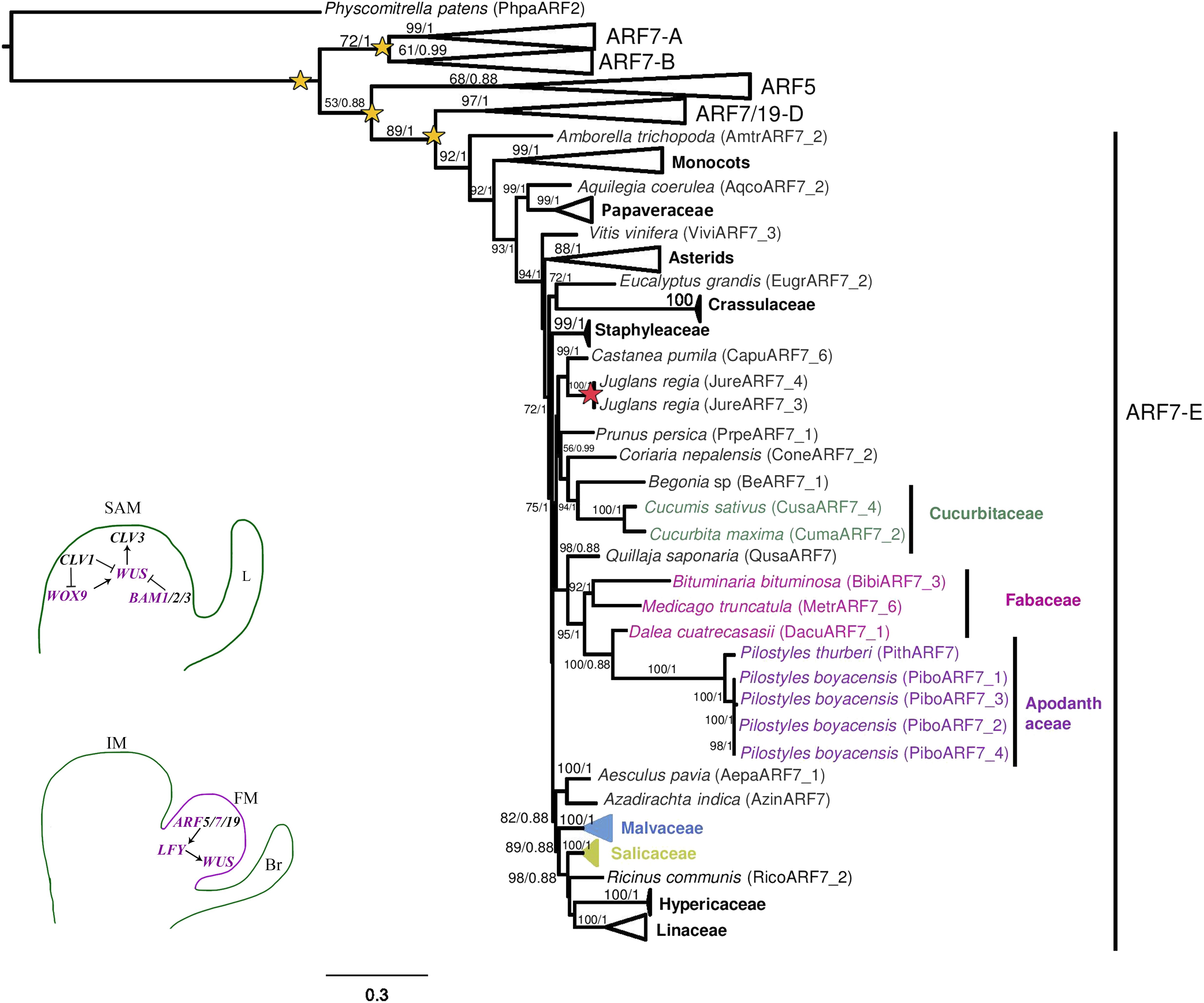
Figure 6. Maximum likelihood reconstruction of the AUXIN RESPONSE FACTOR 5/7/19 gene lineage. Superclades without sequences from Pilostyles boyacensis are collapsed. Bootstrap values >50 and posterior probability are shown above the nodes. Yellow stars indicate large scale or family-level duplication events; red stars indicate species-specific duplication events. Schemes in the bottom left follow the conventions in Figure 2.
Within ARF7-B a large-scale duplication likely occurred in core eudicots because there are two clades recovered containing asterid and rosid (Salicaceae and Malvaceae) representatives. However, one of the two clades also includes homologs from Brassicaceae, Fabaceae, and Cucurbitaceae. In addition, species-specific duplications are found in Argemone mexicana, Austrobaileya scandens, Castanea pumila, Gossypium raymondii, Lonicera japonica, Musa acuminata, Orobanche fasciculata, Papaver setigerum, and Papaver somniferum (Supplementary Figure S7).
The ARF5 clade has remained largely as a single copy, with no large-scale or family-level duplications (Supplementary Figure S7). Thus, well-supported gene clades are recovered for the ARF5 homologs in Amaranthaceae, Brassicaceae, Euphorbiaceae, Fabaceae, Linaceae, Malvaceae, Orobanchaceae, Papaveraceae, Poaceae, Salicaceae, and Solanaceae (Supplementary Figure S7). Species-specific duplications are found only in Amaranthus hypochondriacus, Conopholis americana, Gossypium raimondii, Mimulus guttatus, Musa acuminata, and Orobanche fasciculata (Supplementary Figure S7).
In the ARF7-D clade, two family-level duplications can be detected, one in the Brassicaceae resulting in the ARF7 and ARF19 subclades and a second one within Fabaceae. The remaining ARF7-D homologs are retained as single copy except in Amaranthus hypochondriacus, Argemone mexicana, Atropa belladonna, Castanea pumila, Edgeworthia papyrifera, Gossypium raimondi, Hypericum perforatum, Juglans regia, Linum perenne, Mimulus guttatus, and Papaver somniferum (Supplementary Figure S7).
Finally, in the ARF7-E clade, family-level duplications were detected in Poaceae and Solanaceae, but most other homologs remain as single copy. Species-specific duplications were found in Argemone mexicana, Conopholis americana, Hypericum perforatum, Linum perenne, L. usitatissimum, Musa acuminata, Papaver setigerum, P. somniferum, Salix purpurea, and Staphylea trifolia (Supplementary Figure S7). In this clade, four variants were isolated from Pilostyles boyacensis. PiboARF7_2 was found in all three transcriptomes (PbE + D, PbFl, and PbFr), while the variant PiboARF7_1 is only found in the PbFl transcriptome, and variants PiboARF7_3 and PiboARF7_4 were isolated from the PbFr transcriptome. The four P. boyacensis and the single P. thurberi homologs clustered together (BS/PP, 100/1). The BLASTN comparison showed a similarity percentage of 78% and coverage of 55–74% with homologs from Euphorbiaceae and Salicaceae. Additionally, an ARF7 homolog isolated from the PbE + D transcriptome, showing an identity of 85% and coverage of 98% with other species of Fabaceae, was assigned here to the host D. cuatrecasasii and labeled as DacuARF7_1 (Figure 6 and Supplementary Figure S7).
The MEME analysis detected the N-terminal B3 DNA binding and the Auxin response factor domains, as well as the C-terminal AUX/IAA domain (with III-IV motifs) and two motifs with QSL-rich regions (Supplementary Figure S8). Pilostyles proteins present all of these domains, except for one of two QSL-rich regions (Supplementary Figure S8, purple domain; Table 2). No motifs were found to be exclusive for Apodanthaceae, Cucurbitaceae, or Fabaceae proteins.
The LEAFY Gene Family
Finally, the phylogenetic reconstruction of LEAFY genes showed that they are retained as single copy in most species, except in Brassica rapa, Cladrastis lutea, Cucurbita maxima, Kalanchoe laxiflora, Linum perenne, L. usitatissimum, and Zea mays (Figure 7 and Supplementary Figure S9). Three different variants were isolated from Pilostyles boyacensis. PiboLFY1 and PiboLFY2 were found in the PbFl and PbFr transcriptomes, whereas PiboLFY3 was detected only in the PbE + D transcriptome. PiboLFY2 has an additional exon of 135 nt in comparison to PiboLFY1. PiboLFY3 is a partial sequence that differs in two nucleotides located in the 5′UTR (Supplementary Figure S10).
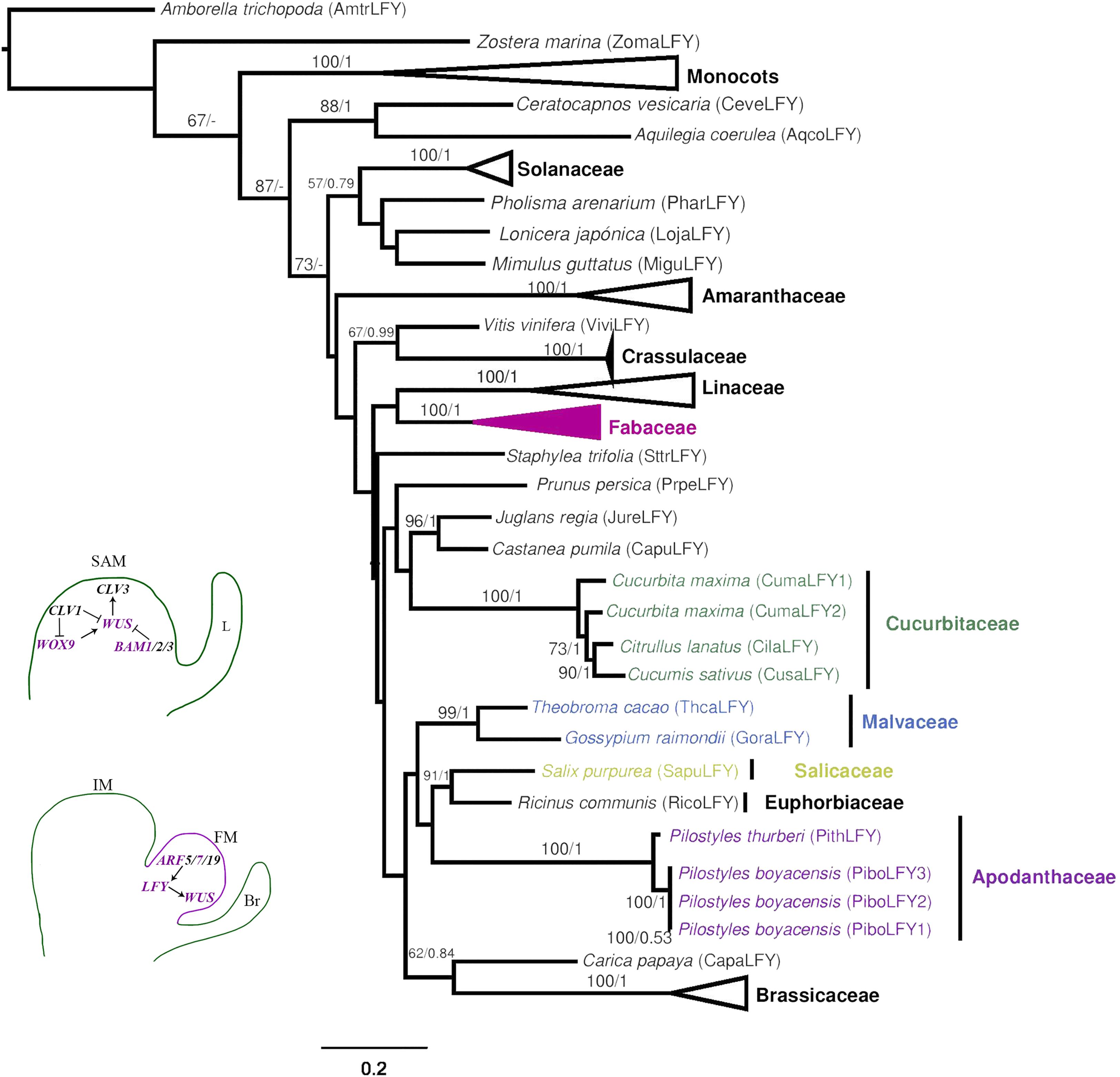
Figure 7. Maximum likelihood reconstruction of the LEAFY gene lineage. Schemes in the bottom left follow the conventions in Figure 2.
The LFY variants from Pilostyles boyacensis cluster with the single copy found in P. thurberi (PithLFY, BS/PP, 100/1, Figure 7). Interestingly, the ML analysis shows Pilostyles homologs as closely related to Euphorbiaceae and Salicaceae genes (BS:38, Supplementary Figure S9), whereas in the BI analysis, they cluster with Cucurbitaceae homologs (PP: 0.78, Supplementary Figure S9). The MEME analysis detects the highly conserved N-terminal Floricaula/Leafy protein SAM domain and the C-terminal DNA Binding Domain Leafy/F. Additionally, an N-terminal LPPP and a GLSEE motif were detected in the full and the reduced datasets (Supplementary Figure 10). Sequences from Pilostyles showed all of these domains and motifs, except for the variant PiboLFY3, which is an incomplete CDS (Table 2). No motifs are exclusive for Apodanthaceae or Cucurbitales sequences.
Discussion
Unlike the typical vegetative SAM to IM to FM developmental sequence that occurs in most angiosperms including Arabidopsis (Ó’Maoiléidigh et al., 2014), the transit to FMs in Pilostyles takes place directly from non-specialized endophytic tissue without recognizable vegetative or IMs (Amaral, 2007; González and Pabón-Mora, 2017). Five out of the 11 A. thaliana canonical gene families responsible for SAM maintenance and floral fate determinacy targeted in this study were found in Pilostyles boyacensis.
The WUSCHEL and WUSCHEL-Related Homeobox 9 Gene Subfamilies
The WUSCHEL-related homologs are represented in Pilostyles boyacensis by PiboWUS, and PiboWOX9. WOX9 homologs have key roles in early embryo patterning, as well as pluripotency and cell proliferation in the SAM across seed plants (Haecker et al., 2004; Nardmann et al., 2007; Palovaara et al., 2010; Skylar et al., 2010; Zhou et al., 2018). In addition, WOX9 homologs also play roles in inflorescence architecture and floral identity in petunia and tomato as well as in carpel development in Arabidopsis, and possibly in sex determination in Cucumis (Wu et al., 2005; Rebocho et al., 2008; Costanzo et al., 2014; Pawełkowicz et al., 2019). PiboWOX9 transcripts were found in all three transcriptomes including both early and late developmental stages. Based on the putative roles previously identified in model taxa, we hypothesize that PiboWOX9 variants have pleiotropic roles in inflorescence architecture, floral identity, carpel development, and perhaps sex determination because they were found in the PbE + D and PbFl transcriptomes. Additionally, PiboW9_2 could be involved in early embryo patterning because it was found in the PbFr transcriptome. Finally, because their protein sequences have a complete N-terminal motif MASSN and two C-terminal motifs VFIN and LQxG WOX8 (Deveaux et al., 2008; Palovaara et al., 2010), at least the PiboWOX9_2 and PiboWOX9_3 isoforms are fully functional.
PiboWUS variants were isolated exclusively from the PbFl transcriptome. Most WUS homologs functionally analyzed control stem cell maintenance in the SAM, the RAM, and the FMs (Laux et al., 1996). Additionally, WUS genes are involved in embryo patterning, leaf, perianth, anther, and ovule development (van der Graaff et al., 2009; Tsuda and Hake, 2016; Wu et al., 2019). At least PiboWUS_2 contains the TLxLFP WUS-box and the EAR-like LELxL motif; the latter is crucial in stem cell maintenance because it mediates the repression of target genes (Kieffer et al., 2006; Tsuda and Hake, 2016). Considering that SAMs in P. boyacensis are lacking and that PiboWUS homologs were not found in the PbE + D or the PbFr transcriptomes, we hypothesize that their functions are restricted to the stem cell maintenance in FMs and perhaps in anther and ovule development.
The Leucine-Rich Repeat Receptor-Like Kinase IX—a Subfamily
The LRR-RLK-IX, a subfamily in Pilostyles, is represented by BAM1/2 homologs while CLAVATA1 homologs are absent. Two copies of BAM1/2 homologs were detected in each of the two species of Pilostyles sampled. The copies are found in different transcriptomes, with PiboBAM1 active in PbE + D and PbFr, and PiboBAM2 active in PbFl and PbFr. At least one variant of each copy conserves the extracellular leucine-rich repeat domain, necessary for detection of CLE ligands, as well as the cytoplasmic kinase domains in charge of phosphorylation of target proteins (Supplementary Figure S6; Hazak and Hardtke, 2016; Wu et al., 2018).
Four different functions have been assigned to CLV1/BAM genes in A. thaliana. The first is related to the role of CLV3, which binds CLV1/2 to repress WUSCHEL related genes in the SAM in order to control cell differentiation and organogenesis (Fletcher, 1999; DeYoung et al., 2006; DeYoung and Clark, 2008; Hazak and Hardtke, 2016; Pan et al., 2016). CLV1 is reinforced by BAM homologs as they sequester CLE ligands secreted in the periphery, resulting in the maintenance of pluripotency in the meristematic field (DeYoung and Clark, 2008). The second, involves BAM1 in the quiescent center of the RAM restricting cell proliferation (DeYoung et al., 2006). The third role involves BAM1/2 in the sporogenous cell fate in the anthers and in ovule development (DeYoung et al., 2006; Li et al., 2016). The fourth role is the control of vascular development in roots, shoots, and leaves by BAM3 (Hazak and Hardtke, 2016).
BAM1/2 transcripts from PbFr lack the kinase domain and have premature stop codons (Table 2 and Supplementary Figure S6). It is likely that such incomplete proteins no longer play a role in early embryogenesis, including putative plumule and radicle differentiation. This is further supported by the fact that CLV1 homologs were not found in any of the Pilostyles transcriptomes, and members of the CLE family (CLV3) identified were assigned by similarity to the host Dalea cuatrecasasii. The isolation of complete BAM1/2 proteins from the PbE + D and the PbFr transcriptomes strongly suggests that the primary role of BAM1/2 in Pilostyles is restricted to anther and ovule development. Nonetheless, a secondary role related to the vascular development of the floral extensor in Pilostyles (Amaral, 2007; González and Pabón-Mora, 2017) cannot be ruled out.
The Auxin Response Factor 5/7/19 Subclade
The ARF5/7/19 subclade in Pilostyles is represented only by ARF7E orthologs. Specifically, PiboARF7E_2 was found in all three transcriptomes, whereas PiboARF7E_1 was detected only in the PbFl, and PiboARF7E_3, PiboARF7E_4 were found only in the PbFr transcriptome. Transcripts from Pilostyles (except PiboARFE7_1) carry intact DNA binding domains as well as AUX/IAA and auxin response factor domains. However, only one out of the two characteristic QSL-rich regions is present in all Pilostyles ARF7E proteins. Both, the AUX/IAA domain and the QSL region have been involved in dimerization with other ARFs (Korasick et al., 2014; Chandler, 2016). Thus, sequence analyses indicate that at least three isoforms of PiboARF7 are fully functional. Nevertheless, the lack of ARF7E orthologs in A. thaliana hinders a direct functional comparison with Pilostyles homologs. Other ARF5/7/19 are known to play redundant roles in lateral root formation, shoot regeneration, leaf polarity and expansion, FM determination, fertilization, fruit development, senescence, and organ abscission (Chandler, 2016). Altogether, our data suggests that the most likely functions of the PiboARF7 homologs include FM determination, fertilization, and fruit development. It is possible that PiboARF7E genes are taking on the function of ARF5 in activating LFY.
The LEAFY Gene Family
LFY homologs in Pilostyles species are present as single copy genes, even though in P. boyacensis three variants were isolated. PiboLFY1 and PiboLFY2 are present in the PbFl and PbFr transcriptomes, while PiboLFY3 is active in the PbE + D transcriptome. The canonical C-terminal DNA binding and N-terminal FLORICAULA/LEAFY domains reported for LFY (Maizel et al., 2005) are conserved in two of the three PiboLFY variants, suggesting that they are functional. LFY is an indirect target of FLOWERING LOCUS T (FT), the florigen, and is a master regulator of FM fate because it represses IM identity and promotes floral organ identity genes (Moyroud et al., 2010). Our results point to conserved roles of PiboLFY genes in establishing FM identity and perhaps together with other cofactors activating floral organ identity in Pilostyles.
Conclusion
The endoparasitic lifestyle of Apodanthaceae is accompanied by a direct transition from a highly reduced endophyte to FMs. We used comparative transcriptomics from three developmental stages of Pilostyles boyacensis with and without the host tissues to investigate the genetic regulatory networks involved in SAM and FM identity and maintenance. Eleven canonical gene families responsible for SAM maintenance and floral fate determinacy were targeted in this study. Only five of them were found in Pilostyles, namely ARF7, BAM1, LFY, WOX9, and WUS. No orthologs of ARF5, ARF19, BAM2, BAM3, CLV1, or CLV3 were found. The lack of SAMs and leaves seems to correlate with reductions in the number of genes for the most important gene families controlling the fine-tuning between pluripotency and organogenesis during vegetative growth. Most genes retained are likely to be functioning in establishing reproductive transition and FM fate, as well as early embryogenesis. We provide the first comprehensive comparative platform to assess meristematic activity in the reduced vegetative parts of an endo-holoparasitic plant.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank under accession numbers MN946521–MN946537. Alignments used are in Supplementary Material S11.
Author Contributions
AG, NP-M, and FG conceived and designed the research and performed field and lab procedures, as well as gene evolution analyses. JA performed the assembly and BLAST analyses. All authors wrote, revised, and approved the submitted version.
Funding
This project was supported by the Universidad Nacional de Colombia, Vicerrectoría de Investigaciones through the Convocatoria Nacional de Proyectos para el Fortalecimiento de la Investigación, Creación e Innovación de la Universidad Nacional De Colombia 2016–2018, project number 37247, and by Universidad de Antioquia, Estrategia de Sostenibilidad 2018–2019 and Convocatoria Programáticas 2017-16302 grants.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the students Sebastián Guzmán and Paola Piñeros (Universidad Nacional de Colombia) for field work support. We also thank Sebastian González (Massachusetts College of Art and Design) for taking the photographs in Figure 1. Preliminary results from this research were presented in the IX Latin American Society for Developmental Biology Meeting (Medellín, Colombia, 2017).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00209/full#supplementary-material
Footnotes
- ^ https://www.illumina.com/
- ^ https://www.hardwoodgenomics.org
- ^ https://github.com/TransDecoder/TransDecoder/wiki
- ^ https://mafft.cbrc.jp/alignment/server/merge.html
- ^ http://meme-suite.org/tools/meme
References
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Čech, M., et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Amaral, M. M. (2007). A estrutura da angiosperma endoparasita Pilostyles ulei (Apodanthaceae): Interface e impacto no lenho de Mimosa spp. Available online at: http://www.teses.usp.br/teses/disponiveis/41/41132/tde-05112007-095018/ (accessed March 11, 2017).
Amaral, M. M., and Ceccantini, G. (2011). The endoparasite Pilostyles ulei (Apodanthaceae – Cucurbitales) influences wood structure in three host species of Mimosa. IAWA J. 32, 1–13. doi: 10.1163/22941932-90000038
Arias-Agudelo, L. M., González, F., Isaza, J. P., Alzate, J. F., and Pabón-Mora, N. (2019). Plastome reduction and gene content in New World Pilostyles (Apodanthaceae) unveils high similarities to African and Australian congeners. Mol. Phylogenet. Evol. 135, 193–202. doi: 10.1016/j.ympev.2019.03.014
Bellot, S., and Renner, S. S. (2014). The systematics of the worldwide endoparasite family Apodanthaceae (Cucurbitales), with a key, a map, and color photos of most species. PhytoKeys 36, 41–57. doi: 10.3897/phytokeys.36.7385
Blarer, A., Nickrent, D. L., and Endress, P. K. (2004). Comparative floral structure and systematics in Apodanthaceae (Rafflesiales). Plant Syst. Evol. 245, 119–142. doi: 10.1007/s00606-003-0090-2
Brasil, B. D. A. (2010). Ciclo de Vida, Fenologia e Anatomia Floral de Pilostyles (Apodanthaceae - Rafflesiaceae s.l.): Subsídios Para um Posicionamento Filogenético da Família Apodanthaceae. Available online at: http://www.teses.usp.br/teses/disponiveis/41/41132/tde-10122010-105707/ (accessed March 11, 2017).
Bryan, A. C., Obaidi, A., Wierzba, M., and Tax, F. E. (2012). XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 235, 111–122. doi: 10.1007/s00425-011-1489-6
Chandler, J. W. (2016). Auxin response factors. Plant Cell Environ. 39, 1014–1028. doi: 10.1111/pce.12662
Chandler, J. W., and Werr, W. (2015). Cytokinin–auxin crosstalk in cell type specification. Trends Plant Sci. 20, 291–300. doi: 10.1016/j.tplants.2015.02.003
Costanzo, E., Trehin, C., and Vandenbussche, M. (2014). The role of WOX genes in flower development. Ann. Bot. 114, 1545–1553. doi: 10.1093/aob/mcu123
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., and Flouri, T. (2019). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294. doi: 10.1093/molbev/msz189
Denay, G., Chahtane, H., Tichtinsky, G., and Parcy, F. (2017). A flower is born: an update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 35, 15–22. doi: 10.1016/j.pbi.2016.09.003
Deveaux, Y., Toffano-Nioche, C., Claisse, G., Thareau, V., Morin, H., Laufs, P., et al. (2008). Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 8:291. doi: 10.1186/1471-2148-8-291
DeYoung, B. J., Bickle, K. L., Schrage, K. J., Muskett, P., Patel, K., and Clark, S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis: BAM receptor kinases regulate meristem function. Plant J. 45, 1–16. doi: 10.1111/j.1365-313X.2005.02592.x
DeYoung, B. J., and Clark, S. E. (2008). BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180, 895–904. doi: 10.1534/genetics.108.091108
Finet, C., Berne-Dedieu, A., Scutt, C. P., and Marlétaz, F. (2013). Evolution of the ARF gene family in land plants: old domains, new tricks. Mol. Biol. Evol. 30, 45–56. doi: 10.1093/molbev/mss220
Fletcher, J. C. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Gaillochet, C., and Lohmann, J. U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142, 2237–2249. doi: 10.1242/dev.117614
González, F., and Pabón-Mora, N. (2014a). First reports and generic descriptions of the achlorophyllous holoparasites Apodanthaceae (Cucurbitales) of Colombia. Actualidades Biol. 36, 123–135.
González, F., and Pabón-Mora, N. (2014b). Pilostyles boyacensis a new species of Apodanthaceae (Cucurbitales) from Colombia. Phytotaxa 178:138. doi: 10.11646/phytotaxa.178.2.5
González, F., and Pabón-Mora, N. (2017). Floral development and morphoanatomy in the holoparasitic Pilostyles boyacensis (Apodanthaceae, Cucurbitales) reveal chimeric half-staminate and half-carpellate flowers. Int. J. Plant Sci. 178, 522–536. doi: 10.1086/692505
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Ha, C. M., Jun, J. H., and Fletcher, J. C. (2010). “Chapter Four - Shoot Apical Meristem Form and Function,” in Current Topics in Developmental Biology Plant Development, ed. M. C. P. Timmermans (Cambridge, MA: Academic Press), 103–140. doi: 10.1016/s0070-2153(10)91004-1
Haecker, A., Gross-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., et al. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668. doi: 10.1242/dev.00963
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hardtke, C. S., Ckurshumova, W., Vidaurre, D. P., Singh, S. A., Stamatiou, G., Tiwari, S. B., et al. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089–1100. doi: 10.1242/dev.00925
Hazak, O., and Hardtke, C. S. (2016). CLAVATA 1-type receptors in plant development. EXBOTJ 67, 4827–4833. doi: 10.1093/jxb/erw247
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kieffer, M., Stern, Y., Cook, H., Clerici, E., Maulbetsch, C., Laux, T., et al. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18, 560–573. doi: 10.1105/tpc.105.039107
Korasick, D. A., Westfall, C. S., Lee, S. G., Nanao, M. H., Dumas, R., Hagen, G., et al. (2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. U.S.A. 111, 5427–5432. doi: 10.1073/pnas.1400074111
Laux, T., Mayer, K. F., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Li, H., Shi, Q., Zhang, Z.-B., Zeng, L.-P., Qi, J., and Ma, H. (2016). Evolution of the leucine-rich repeat receptor-like protein kinase gene family: ancestral copy number and functional divergence of BAM1 and BAM2 in Brassicaceae: evolution of the LRR-RLK gene family. J. Syt. Evol. 54, 204–218. doi: 10.1111/jse.12206
Maizel, A., Busch, M. A., Tanahashi, T., Perkovic, J., Kato, M., Hasebe, M., et al. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308, 260–263. doi: 10.1126/science.1108229
Matasci, N., Hung, L.-H., Yan, Z., Carpenter, E. J., Wickett, N. J., Mirarab, S., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3:17. doi: 10.1186/2047-217X-3-17
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), (New Orleans, LA: IEEE), 1–8.
Moyroud, E., Kusters, E., Monniaux, M., Koes, R., and Parcy, F. (2010). LEAFY blossoms. Trends Plant Sci. 15, 346–352. doi: 10.1016/j.tplants.2010.03.007
Nardmann, J., Zimmermann, R., Durantini, D., Kranz, E., and Werr, W. (2007). WOX gene phylogeny in Poaceae: a comparative approach addressing leaf and embryo development. Mol. Biol. Evol. 24, 2474–2484. doi: 10.1093/molbev/msm182
Nickrent, D. L. (2020). Parasitic angiosperms: How often and how many? TAXON 69, 5–27. doi: 10.1002/tax.12195
Nikolov, L. A., Endress, P. K., Sugumaran, M., Sasirat, S., Vessabutr, S., Kramer, E. M., et al. (2013). Developmental origins of the world’s largest flowers, Rafflesiaceae. PNAS 110, 18578–18583. doi: 10.1073/pnas.1310356110
Ó’Maoiléidigh, D. S., Graciet, E., and Wellmer, F. (2014). Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 201, 16–30. doi: 10.1111/nph.12444
Pabón-Mora, N., Ambrose, B. A., and Litt, A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158, 1685–1704. doi: 10.1104/pp.111.192104
Palovaara, J., Hallberg, H., Stasolla, C., and Hakman, I. (2010). Comparative expression pattern analysis of WUSCHEL-related homeobox 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Picea abies. New Phytol. 188, 122–135. doi: 10.1111/j.1469-8137.2010.03336.x
Pan, L., Lv, S., Yang, N., Lv, Y., Liu, Z., Wu, J., et al. (2016). The multifunction of CLAVATA2 in plant development and immunity. Front. Plant Sci. 7:1573. doi: 10.3389/fpls.2016.01573
Pawełkowicz, M., Pryszcz, L., Skarzyńska, A., Wóycicki, R. K., Posyniak, K., Rymuszka, J., et al. (2019). Comparative transcriptome analysis reveals new molecular pathways for cucumber genes related to sex determination. Plant Reprod. 32, 193–216. doi: 10.1007/s00497-019-00362-z
Poole, R. L. (2007). The TAIR database. Methods Mol. Biol. 406, 179–212. doi: 10.1007/978-1-59745-535-0_8
Rambaut, A. (2006). FigTree Tree Figure Drawing Toll version 1.3.1. Institute of Evolutionary Biology, University of Edinburgh. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed January 31, 2013).
Rebocho, A. B., Bliek, M., Kusters, E., Castel, R., Procissi, A., Roobeek, I., et al. (2008). Role of EVERGREEN in the development of the cymose petunia inflorescence. Dev. Cell 15, 437–447. doi: 10.1016/j.devcel.2008.08.007
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rutherford, R. J. (1970). The anatomy and cytology of Pilostyles thurberi Gray (Rafflesiaceae). Aliso 7, 263–288. doi: 10.5642/aliso.19700702.13
Skylar, A., Hong, F., Chory, J., Weigel, D., and Wu, X. (2010). STIMPY mediates cytokinin signaling during shoot meristem establishment in Arabidopsis seedlings. Development 137, 541–549. doi: 10.1242/dev.041426
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Taylor-Teeples, M., Lanctot, A., and Nemhauser, J. L. (2016). As above, so below: auxin’s role in lateral organ development. Dev. Biol. 419, 156–164. doi: 10.1016/j.ydbio.2016.03.020
Tsuda, K., and Hake, S. (2016). “Homeobox transcription factors and the regulation of meristem development and maintenance,” in Plant Transcription Factors, ed. D. H. Gonzalez (Amsterdam: Elsevier), 215–228. doi: 10.1016/b978-0-12-800854-6.00014-2
van der Graaff, E., Laux, T., and Rensing, S. A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10:248. doi: 10.1186/gb-2009-10-12-248
Wagner, D., Sablowski, R. W., and Meyerowitz, E. M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285, 582–584. doi: 10.1126/science.285.5427.582
Wils, C. R., and Kaufmann, K. (2017). Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 95–105. doi: 10.1016/j.bbagrm.2016.07.014
Wu, C.-C., Li, F.-W., and Kramer, E. M. (2019). Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS One 14:e0223521. doi: 10.1371/journal.pone.0223521
Wu, Q., Xu, F., and Jackson, D. (2018). All together now, a magical mystery tour of the maize shoot meristem. Curr. Opin. Plant Biol. 45, 26–35. doi: 10.1016/j.pbi.2018.04.010
Wu, X., Dabi, T., and Weigel, D. (2005). Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 15, 436–440. doi: 10.1016/j.cub.2004.12.079
Yamaguchi, N., Wu, M.-F., Winter, C. M., Berns, M. C., Nole-Wilson, S., Yamaguchi, A., et al. (2013). A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 24, 271–282. doi: 10.1016/j.devcel.2012.12.017
Zheng, Y., Wu, S., Bai, Y., Sun, H., Jiao, C., Guo, S., et al. (2019). Cucurbit Genomics Database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 47, D1128–D1136. doi: 10.1093/nar/gky944
Keywords: AUXIN RESPONSE FACTOR 7, BARELY ANY MERISTEM, holoparasitic plants, LEAFY, shoot apical meristem, WUSCHEL
Citation: González AD, Pabón-Mora N, Alzate JF and González F (2020) Meristem Genes in the Highly Reduced Endoparasitic Pilostyles boyacensis (Apodanthaceae). Front. Ecol. Evol. 8:209. doi: 10.3389/fevo.2020.00209
Received: 15 January 2020; Accepted: 08 June 2020;
Published: 21 July 2020.
Edited by:
Annette Becker, University of Giessen, GermanyReviewed by:
Lachezar A. Nikolov, University of California, Los Angeles, United StatesLuiza Teixeira-Costa, University of São Paulo, Brazil
Copyright © 2020 González, Pabón-Mora, Alzate and González. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalia Pabón-Mora, lucia.pabon@udea.edu.co
 Angie D. González
Angie D. González Natalia Pabón-Mora
Natalia Pabón-Mora Juan F. Alzate
Juan F. Alzate Favio González
Favio González