Population genetic analyses reveal host association and genetically distinct populations of social parasite Solenopsis daguerrei (Hymenoptera: Formicidae)
- 1Department of Entomology and Plant Pathology, Herbert College of Agriculture, Knoxville, TN, United States
- 2Exponent, Menlo Park, CA, United States
- 3Department of Integrative Biology, College of Science, Oregon State University, Corvallis, OR, United States
- 4Fundación Para El Estudio De Especies Invasivas, Hurlingham, Buenos Aires, Argentina
- 5Programa De Pós-Graduação Em Entomologia E Conservação Da Biodiversidade, Universidade Federal Da Grande Dourados, Dourados, MS, Brazil
Inquiline ant social parasites exploit other ant species for their reproductive benefit because they do not possess a worker caste. Due to their relative rarity in nature, the biology and natural history of inquilines are largely unknown. Likewise, not much research exists that details the close relationship between inquilines and their host(s), and how each organism influences the genetic structure of the other. Here, we conducted a comparative population genetics study to assess patterns of genetic structure within and among populations of inquiline Solenopsis daguerrei and its known fire ant hosts, which includes invasive Solenopsis invicta. Using nuclear and mitochondrial markers, we show that four genetically distinct groups of S. daguerrei likely exist, each with different degrees of host association. Consistent with previous inferences of the inquiline lifestyle, we find that inbreeding is common in S. daguerrei, presumably a result of intranidal mating and restricted dispersal. Results from this study, specifically host association patterns, may inform future biological control strategies to mitigate invasive S. invicta populations.
1 Introduction
Parasitism describes a relationship whereby one organism benefits to the other organism’s detriment. Parasites are ubiquitous in nearly every ecosystem and are found across the tree of life (Petrie and Møller, 1991; Durden and Musser, 1994; Glatzel and Geils, 2009; Burgess et al., 2017). The commonality of this lifestyle means very few species are completely free from exploitation by parasites (Lafferty, 2008). Even species with complex, hierarchical societies, such as eusocial ants, wasps, and bees, are susceptible to parasites, including but not limited to fungi, viruses, and even other social insects (Schmid-Hempel, 1998). Examples in the latter category involving a social insect parasitizing another social insect are referred to as social parasites (Buschinger, 1986; Hölldobler and Wilson, 1990; Buschinger, 2009).
Social parasitism has evolved independently at least 61 times in ants, creating a “mosaic” of life histories and behavioral traits that vary among species (Rabeling, 2020). Several traits present in free-living ant colonies have putatively facilitated the repeated, convergent evolution of social parasitism, including polygyny (presence of multiple reproductive queens in a colony), polydomy (spread of a single colony across multiple nests), and territorial behavior (Buschinger, 2009). Many social parasites share life history traits with their hosts, implying that they have a close phylogenetic relationship with and directly evolve from their host or host species complex (Buschinger, 1986). This phenomenon is known as Emery’s Rule and exists in two forms: a social parasite is said to follow the “strict” version of the rule if it is a direct relative (sister group) to its host(s) or follows the “loose” version of the rule if it is a close, but not a direct relative to its host(s) (Emery, 1909).
Ant social parasites are grouped into one of four categories: xenobiosis, temporary parasitism, dulosis, and inquilinism (Wilson, 1971; Buschinger, 1986; Hölldobler and Wilson, 1990; Buschinger, 2009; Rabeling, 2020). Each category exists on a spectrum of host dependence. On one end, ant social parasites do not need their hosts for anything other than living space (xenobiosis), whereas other ant social parasites have lost the ability to form colonies independently and require a host to raise their brood during several stages of their development (temporary and dulotic social parasites). The other end of the spectrum is inquilinism, considered “true parasitism” because parasites do not possess a worker caste and rely solely on their hosts to rear brood. Inquiline social parasites infiltrate a host colony either by chemical insignificance (thinner cuticle) or by mimicking the chemical profile of their host, which they obtain from host colony members or surrounding nest material (Hölldobler and Wilson, 1990; Lenoir et al., 2001). Inquiline queens are smaller than their free-living host queen counterparts, with their size being proportionate to host workers. It is hypothesized that their miniature size can evolve because they do not require substantial fat reserves due to their mode of dependent colony formation (Keller and Passera, 1989; Aron et al., 2004). Once accepted into a colony, inquilines interfere with the host queen’s ovulation and fecundity, either through chemical castration or depriving queens of nutrients, and subsequently lay eggs, which are reared to maturity by unsuspecting host workers (Wilson, 1971; Buschinger, 1986; Hölldobler and Wilson, 1990; Buschinger, 2009; Rabeling, 2020).
The biology of many inquilines is poorly understood, and most published field studies, genetic studies, and collection efforts of inquilines have been based on limited sample sizes. This is not surprising given their small body sizes (they are similar in size to resident host workers, which presents a challenge when attempting to differentiate them in the field) and their relative rarity in nature (Hölldobler and Wilson, 1990; Trontti et al., 2006). Although there is a dearth of studies on inquilines, several have revealed predictable phenotypic and genetic changes driven by their unique lifestyle. Because of their “true parasite” morphology (e.g., specialized mouthparts and stunted wing development), inquiline social parasites are assumed to be relatively poor dispersers and reportedly engage in intranidal matings (Wilson, 1971; Hölldobler and Wilson, 1990). Host preference, or host associations, can also shape the genetic structure of inquiline social parasites. Many inquilines, such as Mycocepurus goeldii, Acromyrmex insinuator, and Pseudoatta argentina, only parasitize a single host, with host shifts suspected to be rare or unlikely (Gallardo, 1916; Hölldobler and Wilson, 1990; Schultz et al., 1998; Bekkevold and Boomsma, 2000; Rabeling and Bacci, 2010; de la Mora et al., 2020). Host association, along with nestmate recognition chemical cues and other traits of the “inquiline syndrome”, can lead to patchily distributed, isolated, and highly inbred inquiline populations (Wilson, 1971; Hölldobler and Wilson, 1990; Lenoir et al., 2001; de la Mora et al., 2020). For example, an earlier study revealed that sexuals of the inquiline ant Plagiolepis xene have limited dispersal capabilities and typically are restricted to hosts in the same colony network (Trontti et al., 2006). However, another study of this same species revealed that the parasite was able to colonize genetically unrelated, but spatially close host nests (Mardulyn et al., 2014). Consequently, most described species of inquilines have considerably reduced geographic ranges and population densities (aggregated colonies), especially in comparison to their hosts (Hölldobler and Wilson, 1990; Cini et al., 2019).
Like many forms of parasitism, inquilinism is presumed to have a substantial negative impact on the longevity of the host colony. For example, there have been documented cases of inquilines, such as Leptothorax kutteri and Plagiolepis xene, engaging in oophagy, or consumption of host eggs (Franks et al., 1990; Passera et al., 2001). A few studies have reported the extent to which an inquiline impacts the fitness and evolutionary trajectory of its host, and these observations have led to the exploration of using inquilines in biological control efforts for problematic or invasive eusocial species (Williams et al., 2003; Witek et al., 2014).
Solenopsis daguerrei (Santschi), previously characterized in genus Labauchena (Bruch, 1930; Santschi, 1930; Borgmeier, 1949), is an inquiline social parasite that parasitizes several fire ant species belonging to the South American Solenopsis saevissima species group, which includes global invader Solenopsis invicta (Santschi, 1930; Briano et al., 1997; Calcaterra et al., 2000). Two colony social forms exist in most of these fire ant species, including all known host species of S. daguerrei, and vary by queen number: monogyne colonies contain a single egg-laying queen, whereas polygyne colonies contain multiple egg-laying queens.
S. daguerrei appears to have a relatively broad geographic range that coincides with the central distribution of its known fire ant hosts, encompassing parts of Argentina, Brazil, and Uruguay (Silveira-Guido et al., 1973; Briano et al., 1997; Pesquero and Penteado-Dias, 2004). The abundance of S. daguerrei in nature is low and patchy, yet this inquiline is considered a candidate for biological control (Briano et al., 1997). Field observations revealed that fire ant populations parasitized by S. daguerrei have lower mound densities, fewer queens in multi-queen colonies, and decreased brood production compared to non-parasitized populations (Calcaterra et al., 1999). Despite these promising field observations, attempts to artificially rear and transfer S. daguerrei to S. invicta colonies collected in the USA have failed, which further highlights our lack of knowledge regarding the natural history, basic biology, and dispersal of this species (Briano et al., 2002). Two potential reasons as to why these experiments failed could be due to genetic variation in host association or cryptic variation within species.
To extend previous research on this species in an effort to better understand their biology, we conducted a comparative population genetics study to assess patterns of genetic variation within and among natural populations of S. daguerrei and its known fire ant hosts. Using a variety of population genetics tools and two types of molecular markers, we aimed to answer three questions: 1) is S. daguerrei a single, genetically distinct lineage with a large geographic distribution or does it comprise multiple, genetically distinct populations or species?, 2) if genetically distinct populations or species are present, do they exhibit some degree of association with either hosts or colony social form?, and 3) can we elucidate the breeding biology of S. daguerrei? This research will not only contribute to our knowledge of co-evolutionary processes between hosts and parasites but also will reveal the extent of host specialization of S. daguerrei, which, in turn, has implications for the success of this species as a potential biological control agent against invasive S. invicta.
2 Materials and methods
2.1 Ant collection
S. daguerrei and its fire ant hosts were collected from a total of nine geographic sites (six sites in Argentina and three in Brazil) from 2005-2008 (Figure 1; Table S1). Shovel excavation was used to find parasitized colonies, which typically contain one to several S. daguerrei dealate queens and numerous, easily identifiable winged parasite females and males. We collected 10-20 female and male alates of S. daguerrei, as well as 0-6 S. daguerrei dealate queens from a total of 270 parasitized colonies in the two areas with higher known occurrence within its native range. We also collected a paired sample containing a similar number of workers and alates from the nearest unparasitized fire ant colony (~194) at all sites except two in Brazil. Mound contents were transferred to fluon-lined plastic trays, and parasite and host individuals were collected with forceps and placed into 95% ethanol.
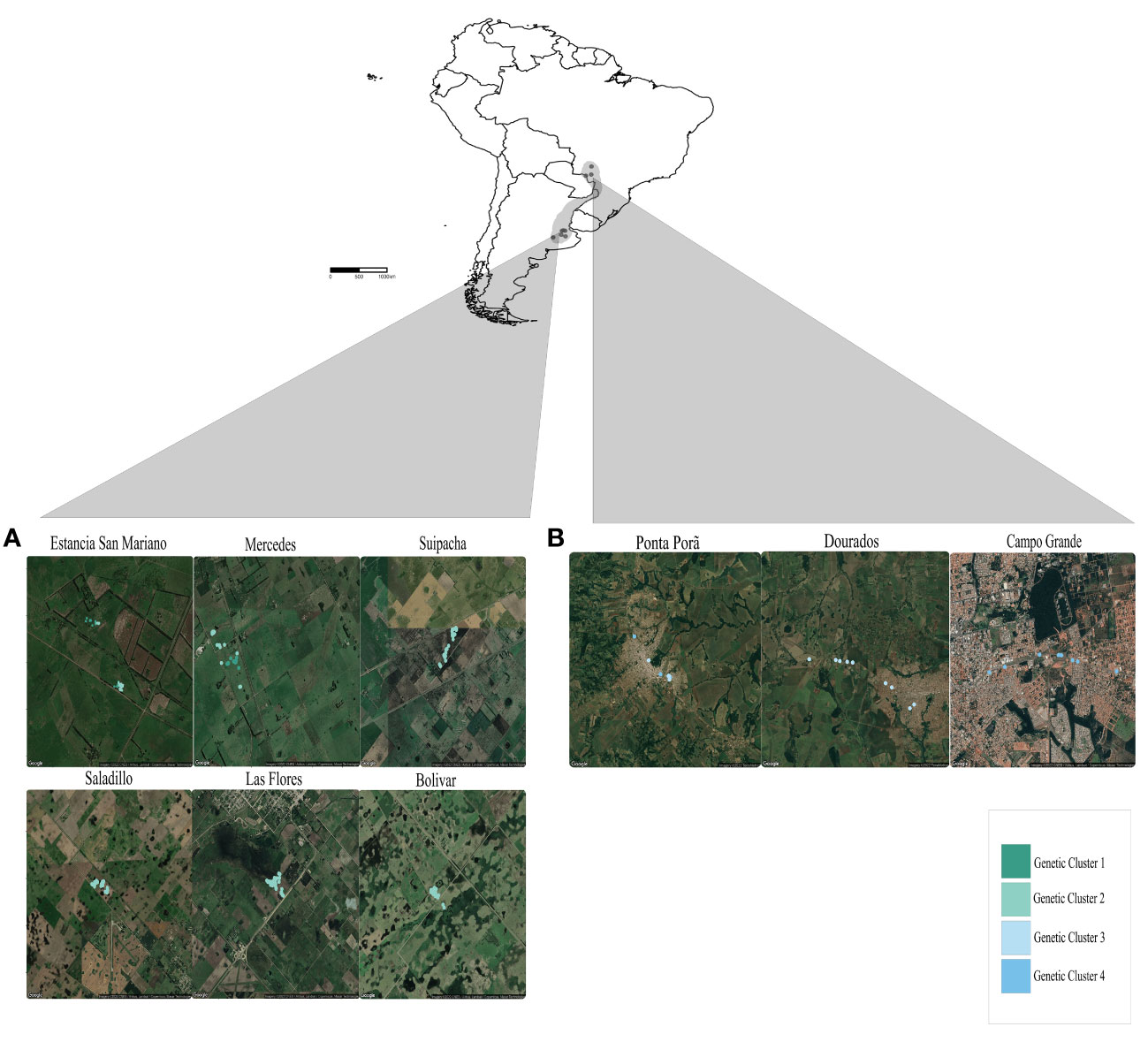
Figure 1 Map of all South American sampling sites of S. daguerrei and fire ant hosts. Shaded area in the country map indicates the putative range of S. daguerrei according to Briano et al. (1997) and Pitts (2002). Insets are satellite maps of all sampled S. daguerrei nests in (A) Argentina and (B) Brazil. The color of each nest corresponds to the genetic cluster assignment from STRUCTURE. Maps were created with R package ggmap (Kahle and Wickham, 2013).
2.2 DNA extractions
Genomic DNA was extracted from one S. daguerrei female alate, one S. daguerrei male, one host female alate from each parasitized colony, and one host female alate from each unparasitized colony using the PureGene DNA Isolation Kit (Gentra Systems) and following the protocol described by Ascunce et al. (2011). DNA extracted from these individuals was used as a template for all downstream PCR and sequencing analyses.
2.3 Microsatellite genotyping
Sixty-six previously developed microsatellite loci were considered for this study (Krieger and Keller, 1997; Chen et al., 2003; Ascunce et al., 2009; Ascunce et al., 2011). Male ants were used to verify the absence of null alleles. Fire ants have a haplodiploid sex determination system, where females are diploid and males are haploid. This unique system allows for easy detection of null alleles using hemizygous individuals, offering a clear advantage over diploid systems and eliminating the need for computer software in this process. Evaluation of missing genotypes for each sample was performed using R package adegenet v.2.1.5 (Jombart et al., 2008). No null alleles were detected in males at any marker. For females, the maximum percentage of missing data was <7%. All samples were retained for downstream analyses.
The final number of microsatellite markers differs between the parasite and host datasets because some loci failed to amplify consistently, and others were monomorphic across all surveyed ants. The final parasite dataset contained 47 variable markers for 270 individuals and the host dataset contained 62 variable markers for 464 individuals (parasitized and non-parasitized nests). A table of microsatellite markers and primer sequences used for this study, adapted from Table S6 of Ascunce et al. (2011), can be found in Table S2.
2.4 MtDNA sequencing
A ~785bp fragment of the cytochrome oxidase subunit I (cox1) mitochondrial gene was amplified via polymerase chain reaction (PCR) using primers C1-J-2195 (5’-TTGATTTTTTGGTCATCCAGAAGT-3’; Simon et al., 1994) and JerryGarcia-C1 (5’-GGGAATTAGAATTTTGAAGAG-3’; Shoemaker et al., 2006). PCR conditions, thermal cycling, and sequencing reactions are outlined in detail in Ahrens et al. (2005). In brief, PCR reactions were carried out in 30-µl volumes containing 8 µl PCR supermix (Denville, Metuchen, NJ), 10.2 µl water, 0.4 µl of each 50-mM primer, and 1 µl genomic DNA under the following cycling profile: 1 min at 94°C for one cycle; 30 sec at 94°C, 1 min at 48°C, 2 min at 68°C for 35 cycles; and 5 min at 72°C for one terminal cycle. Mitochondrial DNA amplicons were purified using Agencourt (Beverly, MA) Ampure magnetic beads and used directly in standard fluorescent cycle-sequencing PCR reactions (ABI Prism Big Dye terminator chemistry; Applied Biosystems, Foster City, CA). Sequencing reactions were purified using Agencourt CleanSEQ magnetic beads and run on an ABI 3700 sequencer. All mtDNA amplicons were sequenced in both directions and corresponding sequence pairs were edited and combined into a single consensus sequence file using the program AutoAssembler (Applied Biosystems).
2.5 Fire ant colony social form
Monogyne and polygyne social form in fire ants is determined by a large non-recombining supergene, which contains hundreds of linked genes that determine many behavioral characteristics that differ between the social forms (Wang et al., 2013). Genotype determination at gene Gp-9, a gene residing within the non-recombining social chromosome, can be used to distinguish colony social form, with monogyne colonies possessing only SB-chromosomes (Gp-9B) and polygyne colonies possessing both SB- and Sb-chromosomes (Gp-9B and Gp-9b; Ross, 1997; Ross and Keller, 1998; Krieger and Ross, 2002). DNA was bulk extracted from ten workers from each parasitized and non-parasitized host colony and used in allelic discrimination assays to identify social form genotype at gene Gp-9. Details of protocols are described in Shoemaker and Ascunce (2010).
2.6 Data analyses
2.6.1 Population structure: clustering methods
The program STRUCTURE v.2.3.4 was used to identify the percentage of cluster membership for all parasite and host individuals (Pritchard et al., 2000). Under the admixture model, STRUCTURE was run separately on the parasite and host (i.e., individuals from both parasitized and non-parasitized fire ant nests) nuclear datasets, with K values ranging from 1 to 10, 500,000 burnin, 1,000,000 MCMC replicates, 30 replicates for each K (300 total), and POPFLAG=0. STRUCTURE tends to detect genetic structure at the highest, most divergent level, with lower-level structure often remaining undetected unless subsequent analyses on partitioned data are performed. To correct this, we performed additional STRUCTURE analyses on the initial clusters obtained for both S. daguerrei and its hosts. Genetic clusters identified by STRUCTURE provide the basis for all subsequent analyses.
The software STRUCTURE assumes genetic markers are unlinked but a previous study in S. daguerrei revealed significant linkage among numerous microsatellite markers, likely attributed to low effective population size and high levels of inbreeding (Ascunce et al., 2009). To address this issue, we ran STRUCTURE on a pruned dataset consisting of 13 unlinked microsatellite markers (one per chromosome, except for chr1, chr9, and chr13). A list of the unlinked loci, along with the genomic and chromosomal positions, can be found in Table S3.
We calculated deltaK with Structure Harvester v.0.6.94 to infer the optimal number of genetic clusters (K) for all datasets (Evanno et al., 2005; Earl and vonHoldt, 2012). Visual inspection of the results using Clumpak (Kopelman et al., 2015) revealed two outlier host ant individuals from Estancia San Mariano and Ponta Porã. These most likely represent fire ant species that are not potential host species and were removed from further analyses given neither were parasitized.
As a model-free supplement to STRUCTURE, we performed a Discriminant Analysis of Principal Components (DAPC) analysis using the R package adegenet v.2.1.5 (Jombart et al., 2008). DAPC, unlike traditional PCA, aims to identify distinct genetic clusters within a dataset by simultaneously maximizing between-group variation and minimizing within-group variation (Jombart et al., 2010). We considered two scenarios for our DAPC analyses of the parasite dataset. The first scenario grouped individuals solely on geographic site assignment, similar to the initial STRUCTURE runs on the combined dataset. The second scenario grouped individuals based on the genetic cluster assignments obtained from STRUCTURE.
2.6.2 Breeding biology: departures from HWE and isolation by distance
Summary statistics, such as observed (HO) and expected (HE) heterozygosities and inbreeding coefficients (FIS) were calculated within and among each parasite and host genetic cluster using hierfstat v.0.5.11 and poppr v.2.9.3 (Goudet, 2005; Kamvar et al., 2014). We performed these calculations on each of the identified genetic clusters (abbreviated throughout as “GC1-4”), irrespective of sampling location. Since all clusters spanned multiple sites, we also performed these calculations for every cluster at each sampling site (e.g., GC1 from Mercedes). In addition, we performed a classical chi-squared tests (hw.test function; pegas package v.1.1 (Paradis, 2010)) to compare the observed genotypic distributions for each marker within and among genetic clusters to those expected under Hardy-Weinberg equilibrium (HWE). For markers out of HWE, we recorded whether a given departure was due to an excess or deficiency of heterozygotes. Any genetic cluster at a given site with less than 10 individuals and any genetic markers with a minor allele frequency of less than 5%, including invariant markers, were excluded from the analysis.
We performed a Fisher’s exact test with Bonferroni correction within and among genetic clusters to assess whether S. daguerrei preferentially parasitizes polygyne host colonies. For our analysis within genetic clusters (e.g., a single genetic cluster spanning several sampling sites), we only used GC2 for two reasons: 1) GC2 had the largest sample size among all clusters (N=201) and 2) we had data for unparasitized colonies at all six sampling sites where GC2 was present (sites 1-6).
Field observations of S. daguerrei sexuals departing from nests suggest restricted dispersal limited mostly to neighboring colonies, which supports previous assumptions of restricted inquiline movement (Hölldobler and Wilson, 1990; Calcaterra et al., 2001). Isolation by distance was measured using Mantel tests (the correlation between genetic and geographic distance) for each parasite and host genetic cluster partitioned by sampling site using vegan v.2.5.7 (Oksanen et al., 2020). Meirmans (2015) reported that solely relying on Mantel tests is insufficient to diagnose isolation by distance among individuals in a population. As an additional test, we performed a standard redundancy analysis (RDA) with Genodive v3.06 (Meirmans, 2020) to determine if spatial distribution has an influence on genetic variation for each parasite and host genetic cluster. Total explained genetic variance was calculated by multiplying constrained variance by overall G’ST for each genetic cluster (Meirmans, 2015).
2.6.3 Breeding biology: clonality
In an attempt to determine if S. daguerrei reproduces clonally, we used the poppr package to calculate the number of multilocus genotypes (MLGs) for each genetic cluster as a whole and each cluster at each sampling site. The ratio between the number of multilocus genotypes and the number of individuals in a population (G/N) may vary with alternate modes of reproduction, with values close to zero suggesting clonal reproduction and values close to one suggesting sexual reproduction (Brunton et al., 2021).
Additionally, we used Genodive v3.06 (Meirmans, 2020) to test for clonality within S. daguerrei. Assignment of clones and evaluation of clonal population structure was based on the stepwise mutation model and corrected Nei’s genetic diversity index, with missing data excluded. The threshold, or the maximum distance allowed between two individuals to assign them to the same clonal lineage, was determined using the methods outlined by Meirmans and van Tienderen (2004). Significance testing to determine if the observed clonal diversity is due to random mating or clonality was performed on each genetic cluster as a whole and partitioned by sampling site.
2.6.4 Genetic divergence and phylogenetic relationships within and among parasites and hosts
Genodive v3.06 (Meirmans, 2020) was used to calculate pairwise G’ST to determine the degree of similarity within and among parasite and host clusters. G’ST, a variant of the traditional FST estimator, is standardized based on the level of population genetic variation and takes into account the multiallelic nature of microsatellites (Hedrick, 2005). We also used the R package haplotypes v.1.1.2 to calculate analogous pairwise ɸST for the mtDNA sequence data (Aktas, 2020). Each pairwise comparison for both genetic differentiation measures were subjected to significance testing using 999 permutations. Pairwise G’ST and ɸST values were estimated between GCs from the same site (sympatric: e.g., GC1 vs GC2 from Mercedes), between GCs from different sites (allopatric: e.g., GC1 from Estancia San Mariano vs GC3 from Ponta Porã), and within GCs between sites (allopatric: e.g., GC3 from Ponta Porã vs GC3 from Campo Grande).
A Bayesian inference tree of S. daguerrei and host mtDNA haplotypes was constructed with MrBayes v.3.2.2 (Huelsenbeck and Ronquist, 2001) using the CIPRES Science Gateway (Miller et al., 2010). We used Mesquite v.3.6.1 (Maddison and Maddison, 2019) to create the input NEXUS file, which included new and previously published S. daguerrei haplotype sequences, as well as sequences from known S. daguerrei hosts, S. pusiliginus, S. electra, S. geminata, and S. amblychila, with the latter two serving as the outgroup (Ross et al., 2003; Ross and Shoemaker, 2005; Shoemaker et al., 2006; Gotzek et al., 2015). We used MrModelTest v.2.4 (Nylander, 2004) and the Akaike Information Criterion (AIC) to select the best substitution model. The tree was generated using default parameters and an MCMC chain of 10,000,000 generations, with data sampled every 1,000 generations, burnin set to discard 25% of samples, and convergence verified by the plot of generation vs log probability of the data. As a supplement to the Bayesian tree, we also constructed a Maximum Likelihood (ML) tree using IQ-Tree v.1.6.12 (Nguyen et al., 2015) with bootstrap support values generated from 1000 replicates under the TPM2u+F+I substitution model, selected with ModelFinder (Kalyaanamoorthy et al., 2017).
3 Results
3.1 Population structure: clustering methods
Our STRUCTURE analyses revealed hierarchical genetic structure within S. daguerrei and its hosts. Considering the data for S. daguerrei only, the number of inferred clusters for the full dataset (Argentina+Brazil) was K=2, with one genetic cluster comprising all individuals from the six sites in Argentina and the other cluster comprising all individuals from three sites in Brazil (Figures S1, S2). The deltaK plots and Clumpak visualizations revealed further substructure; subsequent STRUCTURE runs on these identified clusters resulted in K=4; all individuals comprising two of the four clusters (GC1 and GC2) were confined to Argentina, and individuals in the two remaining clusters (GC3 and GC4) were confined to Brazil (Figure 2A). Out of the nine total sites, six sites comprised individuals from only one of the four genetic clusters (Suipacha (GC2), Saladillo (GC2), Las Flores (GC2), Bolívar (GC2), Dourados (GC4), and Campo Grande (GC3)). The remaining three sites had two distinct parasite clusters as follows: Estancia San Mariano (GC1 and GC2), Mercedes (GC1 and GC2), and Ponta Porã (GC3 and GC4). The average coefficient of membership for all individuals from Argentina is 0.99 (range from 0.73 to 0.99), indicating near-perfect membership to a single genetic cluster; similar coefficients of membership are also seen in the Brazil parasite clusters (Table S4). These results suggest very limited or no gene flow among clusters.
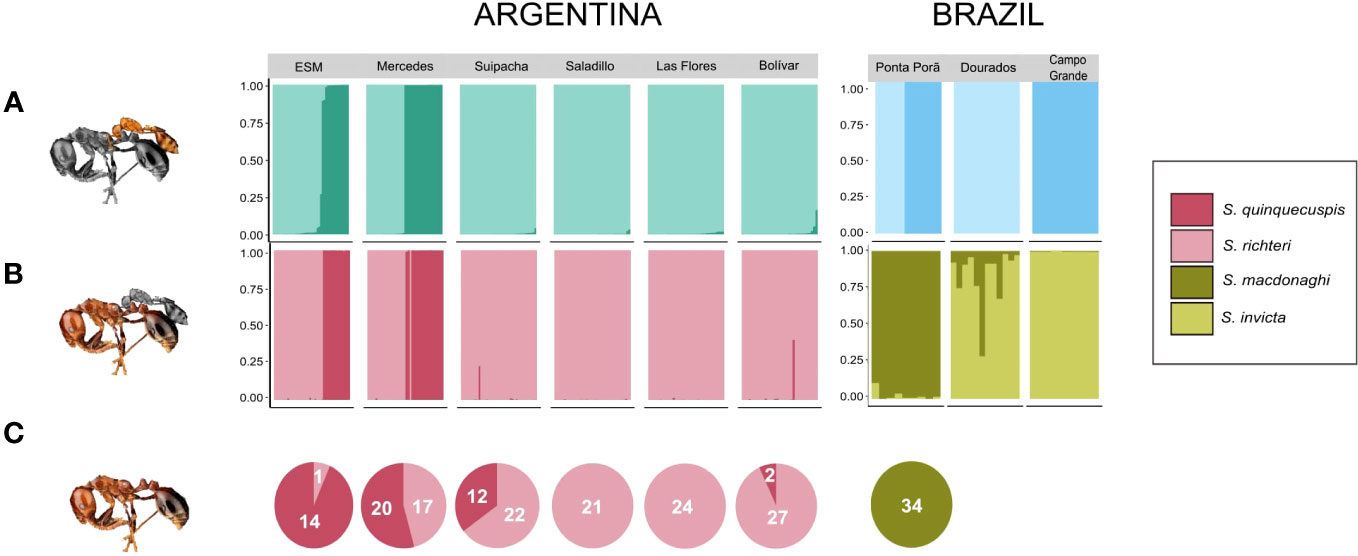
Figure 2 STRUCTURE plots for (A) S. daguerrei and (B) respective hosts across the sampled range. Parasite and host datasets each revealed K=4, with each color representing a distinct genetic cluster. Key matches putative host species to the genetic clusters obtained by the program. Number of unparasitized nests of each host species (C) were reported for most sampling sites, except Dourados and Campo Grande in Brazil.
After visual inspection and additional hierarchical runs, the STRUCTURE results for the pruned S. daguerrei dataset were nearly identical to that of the full dataset, resulting ultimately in the same number of inferred genetic clusters (K=4) and identical assignments of individuals to each of these distinct clusters. The initial STRUCTURE analysis on the pruned dataset (13 microsatellites; Argentina+Brazil) revealed K=3 as the number of inferred clusters, which closely resembled the clustering seen in the full dataset after visual inspection and additional STRUCTURE runs (K=4). However, individuals from Estancia San Mariano and Mercedes (previously GC1 in the full dataset) grouped in a single cluster with individuals from Dourados and Ponta Porã (previously GC4 in the full dataset) (Figure S2; purple cluster in Figure S3). An additional STRUCTURE analysis on this cluster unveiled further substructure, resulting in K=2, in which individuals from Estancia San Mariano and Mercedes were grouped into one cluster, while individuals from Dourados and Ponta Porã were grouped into another. The hierarchical analysis revealed four genetic clusters for the pruned dataset, thus recovering the patterns observed in the full dataset.
We also ran STRUCTURE analyses for the hosts, employing the above hierarchical scheme for both parasitized and non-parasitized hosts. Remarkably, the overall patterns observed in the hosts are identical to those in the parasite. The optimal number of genetic clusters was two (K=2) for the full dataset, with all individuals from Argentina belonging to one cluster and all individuals from Brazil belonging to another (Figure S4). Subsequent STRUCTURE analyses on identified host clusters also revealed four genetic clusters (K=4). Each genetic cluster corresponds to a known host species of S. daguerrei in the S. saevissima species group (GC1: S. quinquecuspis, GC2: S. richteri, GC3: S. macdonaghi, GC4: S. invicta). S. quinquecuspis and S. richteri were only found in sampled sites in Argentina, and S. macdonaghi and S. invicta were only found in sampled sites in Brazil (Figures 2B, C).
A comparison of membership coefficients of parasites and hosts from the same nest indicates a high degree of host association in S. daguerrei. In Argentina, we found near complete concordance in cluster assignments between the parasite and hosts (Figures 2A, B). That is, each S. daguerrei individual in a genetic cluster faithfully matches a single but distinct fire ant host, with only one parasite-host pair as an exception in Mercedes, Argentina. In Brazil, concordance between parasites and hosts was lower, with host association varying among sites. Inquilines belonging to either of two distinct parasite clusters (GC3 and GC4) parasitize S. macdonaghi in Ponta Porã. However, parasites belonging to both clusters seemingly parasitize S. invicta at the other two sampling sites, Dourados and Campo Grande.
The results of the DAPC analyses were largely consistent with our STRUCTURE results in that the individuals from Argentina are genetically different from the individuals in Brazil (Figure S5A). A closer inspection of the results indicates a minor separation of parasite individuals from the different Argentina sites and a much higher separation among parasites in each of the Brazil sites (Figure S5A). Grouping individuals based on genetic cluster assignments from STRUCTURE revealed similar clustering patterns (Figure S5B). All individuals in GC1 and GC2, which are confined to Argentina, cluster together, whereas clusters GC3 and GC4 exclusive to Brazil differ from each other and from GC1 and GC2. However, GC4 is widely separated from all clusters, a pattern that was also observed in our other analyses.
3.2 Breeding biology: departures from HWE and isolation by distance
Nearly half of 47 nuclear markers were near or at fixation within parasite clusters, although which markers were invariant varied among clusters. These markers were removed before HWE tests, resulting in the following total number of variable markers for each genetic cluster: 17 (GC1), 20 (GC2), 18 (GC3), and 21 (GC4). The observed genotypic distributions at each variable marker within S. daguerrei differed significantly from those expected under Hardy-Weinberg equilibrium (HWE) for each of the four genetic clusters (e.g., GC1) and within each genetic cluster at every collection site (e.g., GC1 from Mercedes). All but one of these significant departures were due to fewer than expected numbers of heterozygous individuals (Table S5). Overall observed heterozygosities across all variable loci within and among parasite clusters were much lower than expected, and inbreeding coefficients (FIS) were high (0.4253 to 0.6464) (Table 1).
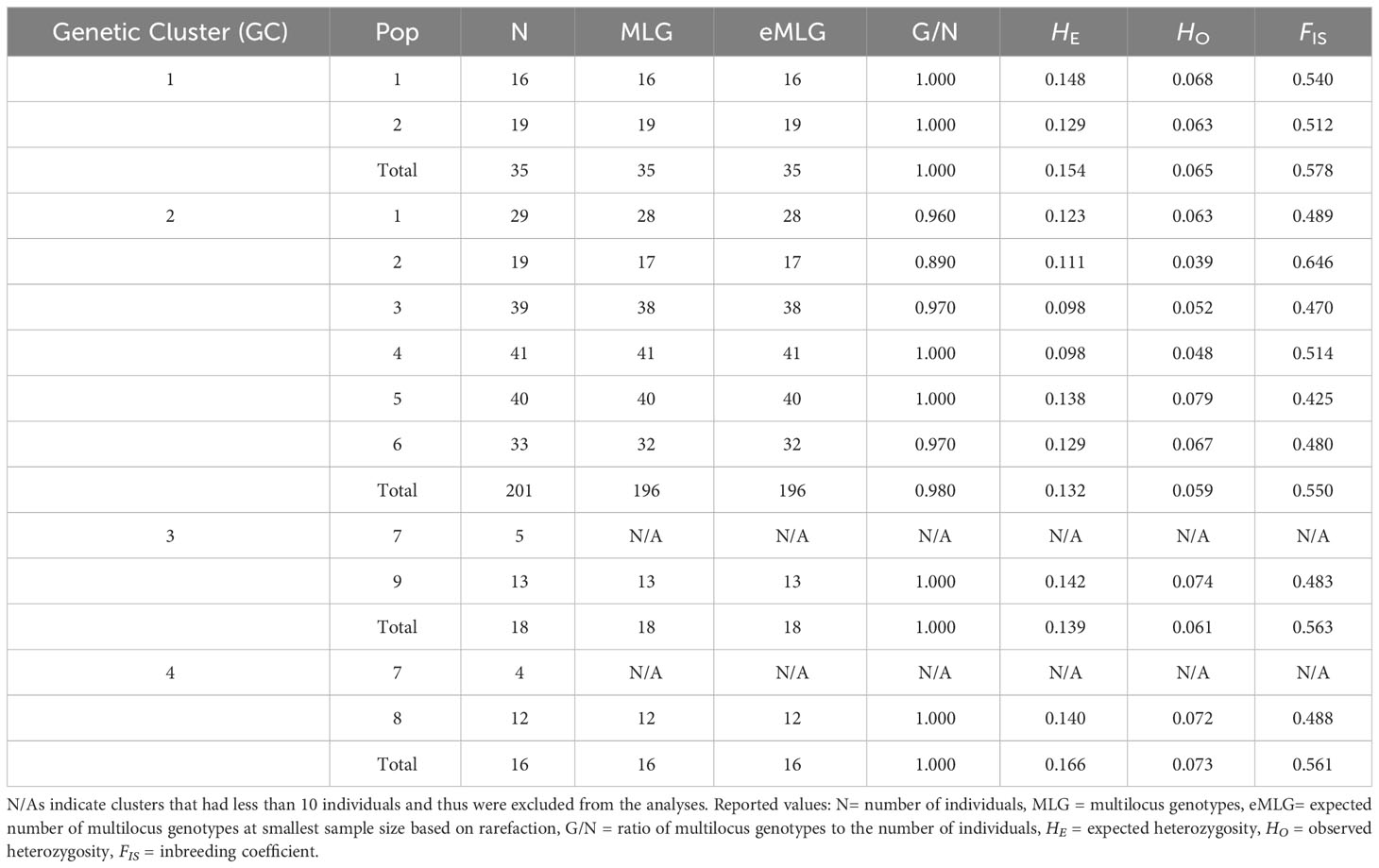
Table 1 Reported summary statistics of each S. daguerrei cluster as a whole and partitioned by sampling site.
In contrast, only 7-13 out of 62 nuclear markers were at or near fixation across all host species, resulting in the following total variable markers: 55 (S. quinquecuspis), 54 (S. richteri), 49 (S. macdonaghi) and 50 (S. invicta). The observed genotypic distributions for many variable markers within the hosts did not significantly differ from those expected under HWE. Further, most of the significant departures were due to heterozygote deficiencies among host species and within each host species at every sampling site (Table S5). However, overall observed heterozygosities across all variable loci within and among species were equal to or slightly lower than expected and inbreeding coefficients were low (-0.0192 to 0.0502), which contrasts sharply with the results for S. daguerrei (Table 2).
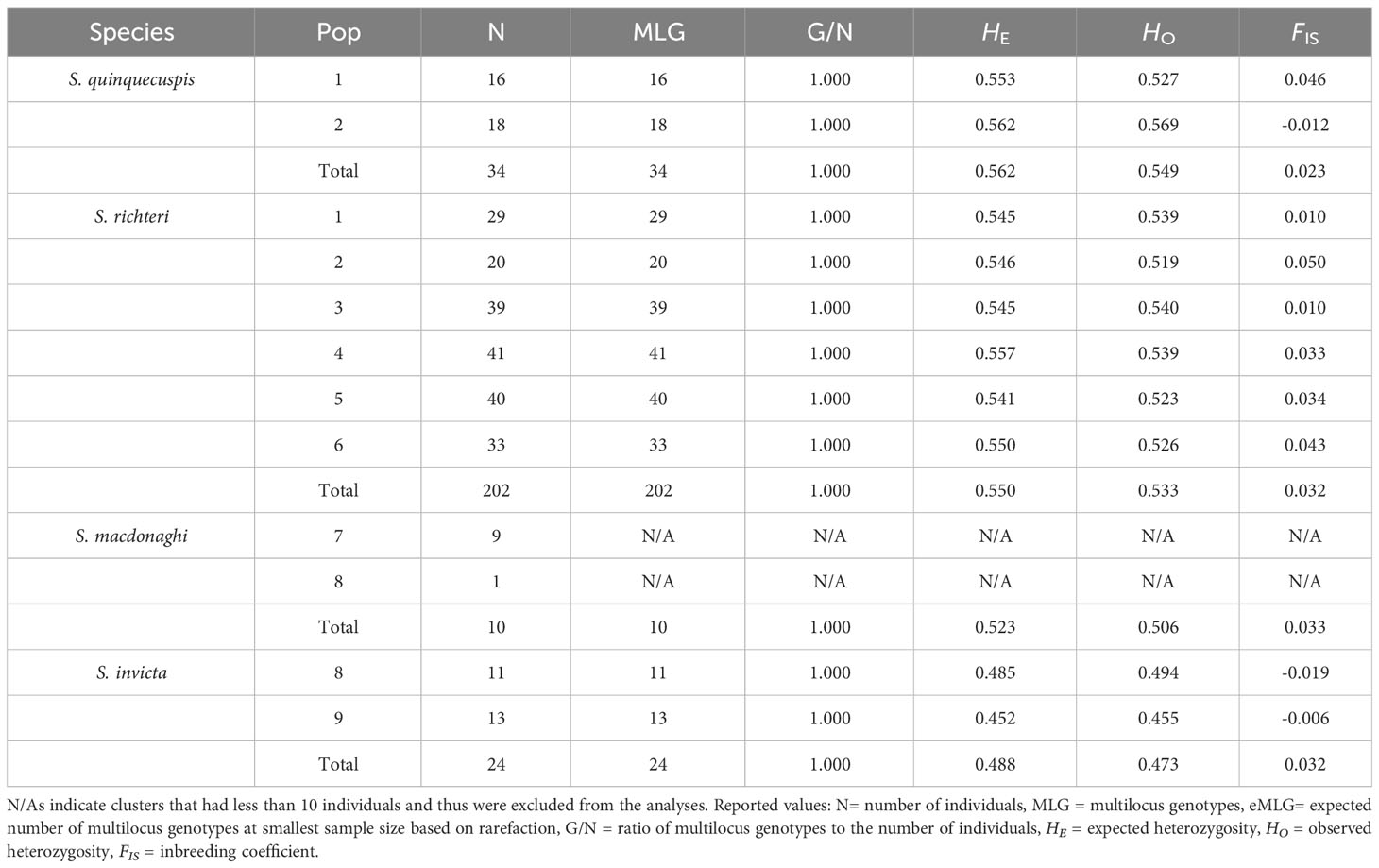
Table 2 Reported summary statistics of each host species as a whole and partitioned by sampling site.
Rates of parasitism in terms of social form were not significantly different within and among clusters. Overall analyses between clusters did not reveal significant non-random associations between parasite presence and social form (p > 0.05 after Bonferroni correction). We also observed no significant associations between parasite presence and social form within any of the six sites in Argentina (p > 0.05 after Bonferroni correction).
Widespread clonality does not seem to be present in S. daguerrei. Comparisons of observed MLGs to the number of individuals within genetic clusters indicate sexual reproduction (G/N close or equal to 1; Table 1). Additionally, the observed clonal diversity within genetic clusters was not significant (attributed to clonal reproduction) in all genetic clusters with the single exception of GC4 (Table S6).
Isolation by distance varied across the genetic clusters. The Mantel tests revealed that significant isolation by distance was seen in individuals of GC1 in Estancia San Mariano and Mercedes, GC2 in Estancia San Mariano, and GC3 in Campo Grande. However, individuals of GC2 across the remaining five sampling sites and all individuals in GC4 do not display significant isolation by distance. Similar results were seen in the host; only populations of S. quinquecuspsis in Estancia San Mariano and Mercedes and S. richteri in Estancia San Mariano experience significant isolation by distance (Table S7). RDA results indicate isolation by distance has little impact on genetic variation; spatial distribution accounted for less than 10% of total genetic variation among the S. daguerrei clusters and even less so among the host clusters (<3%; Table S8).
3.3 Genetic distances
Pairwise G’ST for the parasite and host datasets indicate statistically significant genetic differentiation under all three scenarios (Between GC, Allopatric; Between GC, Sympatric; Within GC, Allopatric) (Figure 3; Table S9). Pairwise G’ST distances between genetic clusters, both in sympatry and allopatry, were larger in S. daguerrei (“Between GC, Sympatric” = 0.47-0.99 and “Between GC, Allopatric” = 0.40-0.99) compared with its respective hosts (“Between GC, Sympatric” = 0.10-0.55 and “Between GC, Allopatric” = 0.12-0.65). Pairwise G’ST distances between sites within each genetic cluster also were larger in the parasite than in the hosts (parasite “Within GC” = 0.09-0.36 vs. host “Within GC” = 0.0-0.15).
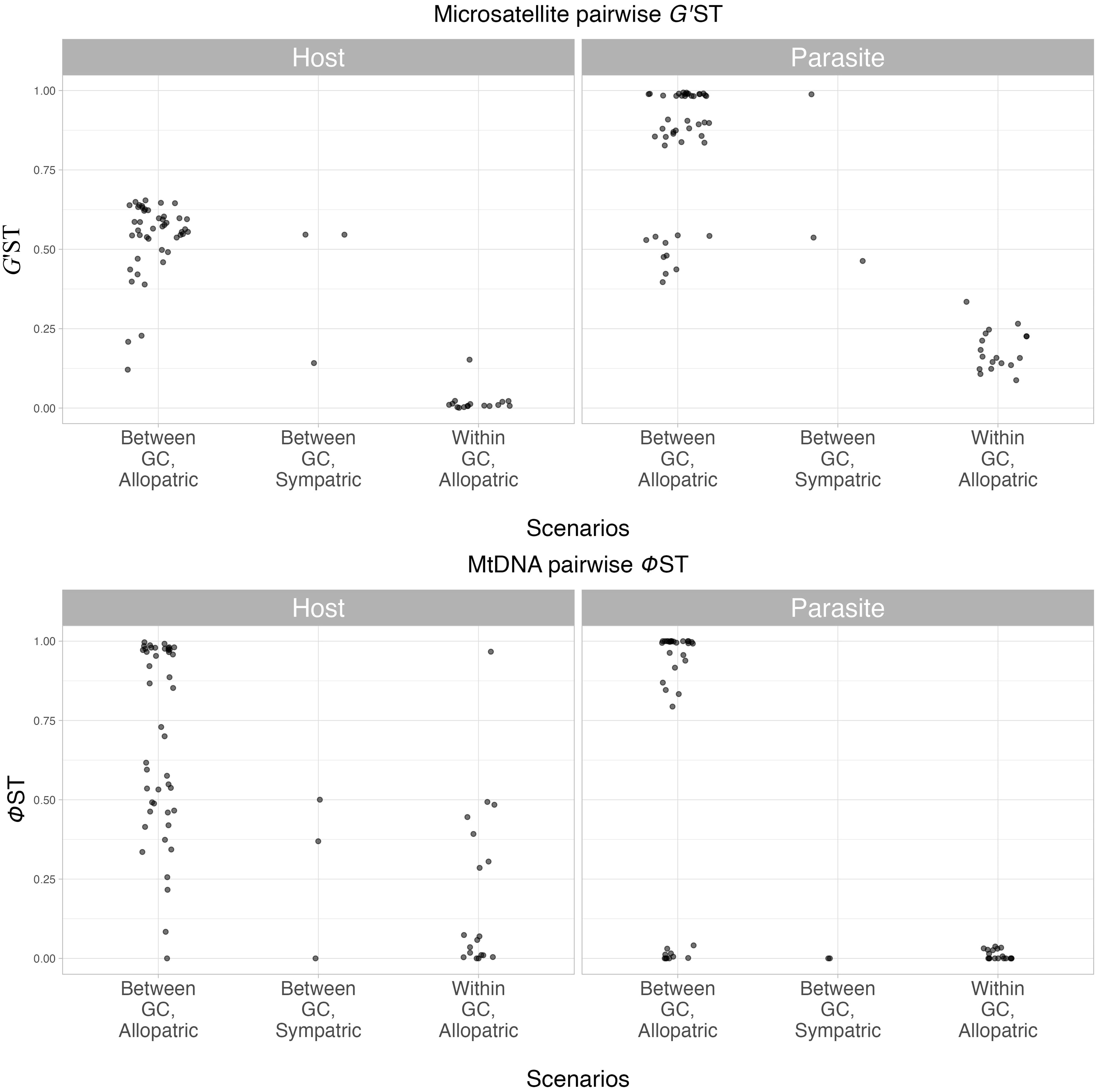
Figure 3 G’ST and ɸST genetic distance comparisons between S. daguerrei and respective hosts. Three scenarios summarize the results: Between GC, Allopatric (between genetic clusters across multiple geographic locations), Between GC, Sympatric (between genetic clusters in the same geographic location), and Within GC, Allopatric (within genetic clusters across multiple geographic locations).
Estimates of pairwise G’ST were consistently high in S. daguerrei, particularly pairwise comparisons involving GC3 and GC4. For example, the pairwise comparison of GC1 in Argentina to GC4 in Brazil revealed highly significant differentiation, with G’ST = 0.98-0.99 (P ≤ 0.001; Table S9A). Pairwise comparisons of G’ST using the pruned dataset were similar to the values using all markers and also highly significant among clusters (Table S9B). Allele frequency comparisons among clusters revealed GC4 has 63 private alleles, a surprisingly large number in comparison to GC1, GC2, and GC3 (Tables S10A, S11A). Private allele counts for the hosts were the largest in GC2, compared with the other GCs (Tables S10B, S11B).
We also estimated the number of cluster-specific fully diagnostic markers for the S. daguerrei clusters. GC4 possessed the largest number of diagnostic markers (Table S10A; Table S12A). GC3 had fewer diagnostic markers when compared with the other clusters, and none of the surveyed genetic markers are completely diagnostic in distinguishing GC1 and GC2. In contrast, we observed very few fully-diagnostic markers for the different host species (Tables S10B, S12B).
Pairwise mitochondrial differentiation (ɸST) varied in both the parasite and the hosts and is especially apparent when compared to nuclear differentiation (Table S13). In the parasite, we observed a wide range in ɸST estimates for comparisons between genetic clusters in both sympatry and allopatry (both ranged from 0.0-1.0). Interestingly, however, the “Within GC, Allopatric’’ scenario revealed considerable discordant patterns between the nuclear and mitochondrial genetic distances. We observed significant nuclear differentiation (G’ST = 0.09-0.39) but much lower mitochondrial differentiation (ɸST = 0.0-0.06). The range in estimates of ɸST in the hosts was large in all three scenarios (“Between GC, allopatric” = 0.02-1.0, “Between GC, sympatric” = 0.0-0.46, and “Within GC, allopatric” = 0.0-0.97). ɸST is only slightly larger than G’ST in the “Between GC, Allopatric” scenario, and larger than G’ST in the “Within GC, Allopatric” scenario, the opposite of the discordant pattern seen in the parasite.
3.4 MtDNA trees
We identified 11 mtDNA haplotypes across all surveyed parasite individuals. Four of these haplotypes were previously published (Shoemaker et al., 2006), whereas the remaining seven are unique to this study. The Akaike Information Criterion (AIC) identified GTR+I as the best substitution model, and the resulting tree depicts two separate clades of S. daguerrei haplotypes (Figure 4). The first clade appears to be monophyletic and contains most of the S. daguerrei haplotypes. Genetic cluster membership for each haplotype, denoted by the pie charts, reveals that many of the haplotypes represent parasites from GC1 and GC2, however, all individuals in GC3 are represented only by a single haplotype (Dag2). The second clade contains one S. daguerrei haplotype (Dag13), along with S. electra and S. pusillignis. Dag13 is exclusive to individuals from GC4 and is highly divergent from the other haplotypes of S. daguerrei. The ML tree had a similar topology to the Bayesian tree, with some weakly supported branches collapsed (bootstrap values less than 80) (Figure S6).
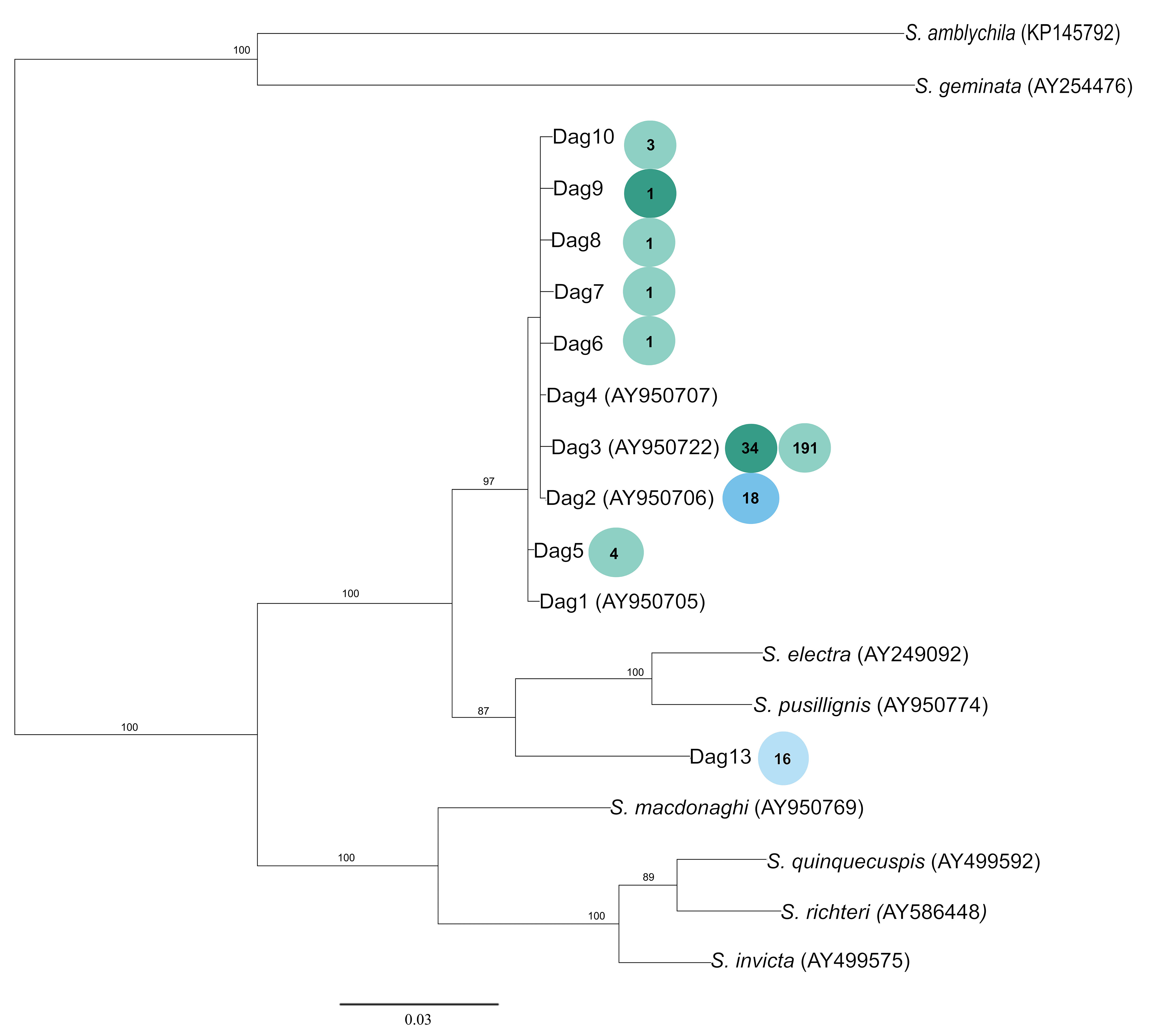
Figure 4 Bayesian mtDNA tree of S. daguerrei and other S. saevissima-species group haplotypes. Previously published sequences are listed with an accession number. Numbers on branches represent the posterior probability (only values greater than 80% are shown). Each circle shows the number of individuals with a given haplotype, with color corresponding to the genetic cluster identified by STRUCTURE (see Figure 2).
4 Discussion
We conducted a detailed population genetics study of the inquiline social parasite Solenopsis daguerrei and its associated host species, with the aims of understanding the influence of geography and host association on shaping inquiline population genetic structure, as well the mating and dispersal biology of S. daguerrei. Three major findings from our study are 1) significant spatial genetic structure exists within S. daguerrei, some portion of which reflects the fact that nominal S. daguerrei is comprised of multiple genetically distinct groups (presumably cryptic species), 2) genetic clusters of S. daguerrei exhibit varying degrees of host association across the sampled range and no preference for social form, and 3) inbreeding and restricted dispersal are common in S. daguerrei, which are likely a result of intranidal mating and are consistent with patterns associated with the inquiline syndrome (Wilson, 1971; Hölldobler and Wilson, 1990; Buschinger, 2009). However, we did not find clear evidence for clonal reproduction in S. daguerrei. We discuss each of these findings below.
4.1 Genetic differentiation within and among Solenopsis daguerrei populations
Our STRUCTURE and DAPC analyses revealed that all surveyed samples of S. daguerrei belong to one of four genetic clusters, which are significantly differentiated from each other. We found six of the nine sampling sites were comprised of individuals from a single genetic cluster (but not the same genetic cluster in all cases) and the remaining three sites (Estancia San Mariano [San Eladio], Mercedes, and Dourados) were comprised of individuals belonging to two distinct genetic clusters. Further, our results suggest gene flow between sympatric clusters at these three sites is limited or absent. For example, all individuals at these three sites exhibit very high membership coefficients to only one of the two clusters, with little to no evidence of admixture (average coefficient of membership to one cluster = 0.99). Pairwise G’ST estimates confirm the sympatric clusters are markedly different from each other (GC1 vs GC2 [ESM] G’ST = 0.47, GC1 vs GC2 [Mercedes] G’ST = 0.54, and GC3 vs GC4 [Ponta Porã] G’ST = 0.99). The large number of diagnostic markers and private alleles for several clusters is also consistent with an absence of significant gene flow.
Additional lines of evidence also indicate S. daguerrei is most likely composed of several closely related species. Aside from the high divergence between sympatric clusters, the “Between GC, Allopatric” scenario shows exceedingly high levels of genetic differentiation among all four parasite clusters, with the highest values (G’ST>=0.80-0.90) involving pairwise comparisons of the other clusters to GC4. Although genetic distances between clusters are higher, comparisons within genetic clusters across geographic locations (“Within GC, Allopatric”) show significant nuclear differentiation (G’ST=0.22-0.40), indicating macrogeographic structure also exists within single allopatric clusters of the parasite, similar to results seen in its hosts (Ross and Trager, 1990; Ahrens and Shoemaker, 2005; Ross and Shoemaker, 2005). Limited or complete lack of ongoing gene flow among genetic clusters (all comparisons except for GC1 and GC2) and considerable divergence from each other suggests S. daguerrei is comprised of multiple genetically distinct groups as opposed to differentiated populations within a single species.
The extreme genetic divergence of GC4 from the other three clusters of S. daguerrei is noteworthy. Our DAPC results reveal that all parasite individuals in GC4 are highly differentiated from individuals belonging to GC1-3. GC4 also contains remarkably high numbers of private alleles (63) and fully diagnostic markers (35-40 markers out of 47 variable markers). Both results, especially the number of fully diagnostic markers, far exceed what we observed among different host fire ant species in this study and earlier studies of fire ants in the S. saevissima species group (Ross and Shoemaker, 2005; Shoemaker, 2016).
The mtDNA haplotype tree also supports the high divergence of GC4 from GC1-3 observed at the nuclear markers. The resulting tree consists of two major clades: one clade consists of individuals from GC1-GC3 and a second clade consists of individuals from GC4, S. electra, and S. pusillignis. GC1 and GC2 share many of the same closely related haplotypes and are not easily distinguishable from each other. All individuals in GC3 have the same mtDNA haplotype (Dag2) that is absent in GC1 and GC2. This mtDNA haplotype (Dag2) falls within a larger clade containing mtDNA haplotypes from GC1 and GC2, but the lack of any shared haplotypes and the absence of gene flow between GC3 and clusters GC1-2 is consistent with early stages of divergence. The second clade or lineage contains haplotypes of S. electra and S. pusillignis, and the single mtDNA haplotype Dag13, which is confined to all individuals of GC4. The high divergence of Dag13 from all other S. daguerrei haplotypes is consistent with our nuclear data suggesting that individuals comprising GC4 represent an entirely different species, potentially previously described but rare inquiline Solenopsis hostilis (Borgmeier, 1959). Very little is known about S. hostilis, as its initial discovery in Rio de Janeiro and Paraná states in Brazil was also the only time it has been collected (Pitts et al., 2005; Pitts et al., 2018).
The pairwise estimates of mtDNA differentiation between S. daguerrei clusters, measured as ɸST, are largely concordant with estimates of G’ST and suggest significant genetic differentiation between inquiline clusters in nearly all scenarios. However, some pairwise ɸST estimates suggest little to no genetic divergence, contrasting a much more differentiated nuclear G’ST, a trend distinctly found in the “Within GC, Allopatric” scenario. Such mitonuclear discordance is prevalent in many systems and a few commonly accepted reasons driving discordance are introgression, sex-biased dispersal, hybrid zone movement, and human mediated habitat disturbance (see review by Toews and Brelsford, 2012). A possible explanation for the incongruence seen among S. daguerrei clusters is it reflects patterns associated with the effects of the bacterial endosymbiont Wolbachia, which are prevalent in many arthropod species, especially fire ants (O’Neill et al., 1992; Shoemaker et al., 2000; Ahrens and Shoemaker, 2005). Many Wolbachia variants are maternally transmitted in insects, and the bacteria have evolved ways to manipulate host reproduction to ensure its transmission (Werren, 1997; Werren et al., 2008). Both theoretical and empirical studies have demonstrated Wolbachia variants that invade new populations may sweep through these populations quickly to high frequencies or fixation if such variants exceed a critical threshold frequency and are favored by selection (Turelli and Hoffmann, 1991; Turelli, 1994). Because these bacteria primarily are maternally transmitted within many species, any selective sweeps of Wolbachia will result in the associated mtDNA haplotype also sweeping to high frequencies because of hitchhiking, such that mtDNA variation within a population is reduced to a single variant and any close mutational derivatives occurring during or after such sweeps (Jiggins, 2003; Tseng et al., 2019; Deng et al., 2021). Some of our results are consistent with patterns associated with a recent selective sweep of Wolbachia in S. daguerrei; the “Within GC, Allopatric” scenario displays a global reduction of mtDNA differentiation, while also exhibiting strong nuclear DNA differentiation. Both marker types are necessary to confirm the presence of a selective sweep, as a maternally linked variant such as Wolbachia has little effect on nuclear variation (Tseng et al., 2019). The scenarios “Between GC, Allopatric” and “Between GC, Sympatric” also display a range of mtDNA differentiation; a small number of pairwise cluster comparisons (comparisons of GC1 to GC2) display low levels of differentiation, while the majority of pairwise cluster comparisons reveal high levels of differentiation (comparisons of GC1 to GC3 and GC4, GC2 to GC3 and GC4, and GC3 to GC4). These patterns could indicate distinct or different combinations of Wolbachia variants exist among the genetic clusters, and that GC1 and GC2 likely share the same variant; this is partially supported by previous research that found up to eight Wolbachia variants in a single S. daguerrei individual (Dedeine et al., 2005).
We also observed mitonuclear discordance within and among the host populations. Significant nuclear differentiation is apparent amongst the host species, but it appears that pairwise mtDNA differentiation is smaller than patterns seen in S. daguerrei, particularly in the “Between GC, Allopatric” scenario. This is consistent with other studies that also found higher mtDNA differentiation among parasites than hosts, as these differences may be attributed in part to the stronger effects of genetic drift in the inquiline parasites as they have smaller effective population sizes than their host counterparts (Bromham and Leys, 2005; Trontti et al., 2005; Trontti et al., 2006; Vepsäläinen et al., 2009). The mtDNA and nuclear DNA markers are largely concordant among the host species, with both having similar levels of differentiation in the “Between GC, Allopatric” and “Between GC, Sympatric” scenarios. However, discordance between the two marker types exists in the “Within GC, Allopatric” scenario in the hosts, where we observe high mtDNA differentiation, but reduced nuclear differentiation, a pattern opposite to the “Within GC, Allopatric” comparisons for the parasite. Instances of mitonuclear discordance that display higher structuring of mtDNA than nuclear DNA can be explained by nuclear introgression and/or sex-biased dispersal (Toews and Brelsford, 2012). This is consistent with previous studies that have revealed similar discordance between the two markers in the fire ant S. invicta; fire ants of the polygyne social form display nuclear homogenization because of male-mediated gene flow, but highly structured mtDNA due to the absence of female-mediated gene flow (Ross and Shoemaker, 1997; Ross et al., 1997).
4.2 Host association of Solenopsis daguerrei
The patterns of genetic differentiation in the inquiline social parasite S. daguerrei are quite striking, especially when aligned with the patterns observed in its hosts. Our nuclear DNA results reveal that in Argentina, all inquiline individuals in GC2 are found only infesting S. richteri, and all inquiline individuals in GC1 are found only infesting S. quinquecuspis, except for a single host-parasite pair in Mercedes. This holds at the two sites where individuals of GC1 and GC2 occur sympatrically. This near-perfect association between the inquiline and its hosts suggests that the inquilines in southern Argentina are highly specialized to a particular host species. The exact mechanisms driving and maintaining host specialization are unknown, and our results could reflect either host association or differential selection and survival due to colonizing hosts across different geographic sites. It is also unclear whether GC1 and GC2 are biologically capable of using other hosts and whether there are fitness consequences for doing so. Fire ants are highly territorial, particularly individuals of the monogyne (one egg-laying queen per colony) social form and will quickly terminate intruders. To avoid detection, S. daguerrei most likely matches its host’s cuticular hydrocarbon profile, a partially heritable chemical system in which ants communicate with each other or fails to be detected due to its thinner cuticle (Hölldobler and Wilson, 1990; Singer, 1998; Tsutsui, 2004). This information, in combination with our results suggesting multiple genetically distinct groups exist within nominal S. daguerrei, may partially explain why previous laboratory attempts at rearing parasites have failed (Briano et al., 2002; Briano et al., 2012).
The patterns of host association seen in S. daguerrei can be reinforced further by examining the non-parasitized host nests in each sampling site. For example, the STRUCTURE plots reveal that the Argentina site Suipacha contains a comparable number of non-parasitized host colonies of both available host species. However, the inquilines in Suipacha unanimously prefer only one host species (GC2 parasitizes nests of S. richteri but not S. quinquecuspis), despite both hosts occupying this site.
Two parasite clusters were found in Brazil, and both are distinct from those in Argentina. However, nuclear evidence for non-random host association is not as clear as it is in Argentina. For example, S. macdonaghi is the only host collected in Ponta Porã and is parasitized by two distinct genetic clusters of S. daguerrei. However, individuals from those same parasite clusters parasitized S. invicta in Dourados and Campo Grande. One potential explanation for this observation is variation in the availability of host species. However, as we lack non-parasitized host data in Dourados and Campo Grande, we can only speculate that the two parasite lineages may opportunistically parasitize multiple hosts in Brazil, and this is partly dependent on host availability at a particular site. A similar trend was seen in another social parasite, the slave-making ant Polyergus mexicanus, where the parasite “makes the best of a bad job” by continuing to remain flexible in its host association (Sapp et al., 2020).
Utilizing a wide range of host species is predicted to impact the diversification of social parasites, which potentially could lead to speciation (Buschinger, 1990; de la Mora et al., 2020). Inquilines, however, are predicted to infrequently engage in host switching due to their extreme specialization primarily on a single host, which may lead to isolated and fragmented populations. This assumption has led others to believe that inquilines are “evolutionary dead ends” (Hölldobler and Wilson, 1990; Jansen et al., 2010; Leppänen et al., 2016; de la Mora et al., 2020); however, in the case of S. daguerrei, we find extensive diversification within the lineage and evidence of colonization of multiple hosts. Future work is needed to assess the evolutionary success of inquilines. Potential research avenues include a comparative genomics approach to detect signatures of gene degradation and gene gains specific to the inquiline lifestyle and investigations into alternative systems of reproduction.
4.3 Solenopsis daguerrei life history and breeding biology
Speciation is believed to occur either due to physical barriers or separation (allopatry) or in the absence of geographical separation within the same area (sympatry) (Coyne and Orr, 2004; Matute and Cooper, 2021). As social parasites and their hosts frequently coexist in the same environment and share many life history traits, it is inferred that they share a close evolutionary relationship. This degree of closeness is described by Emery’s Rule in two versions: the loose form proposes that parasites evolve from their hosts because of physical barriers or isolation (allopatry) and the strict form suggests parasites evolve alongside their hosts in the same geographic area (sympatry) (Emery, 1909). The topology of the mtDNA trees largely indicates a single origin of social parasitism in the S. daguerrei lineage, evidenced by the monophyly formed by the majority of S. daguerrei haplotypes, excluding Dag13. These results corroborate previous studies indicating that S. daguerrei follows the loose interpretation of Emery’s Rule, where the parasite is a non-sister species to and does not cospeciate with its hosts (Pitts et al., 2005; Shoemaker et al., 2006). While S. daguerrei does not seem to evolve in sympatry with its hosts, recent studies have provided evidence of sympatric origins in other ant inquilines in the genera Myrmecia, Formica, and Plagiolepis, further strengthening the understanding of the evolutionary origins of social parasitism in ants (Borowiec et al., 2021; Degueldre et al., 2021; Mera-Rodríguez et al., 2023).
The contrasting modes of inheritance between nuclear and mtDNA markers can yield insight into whether one or both sexes mediate gene flow in S. daguerrei. For example, previous studies revealed that discordant patterns between the two types of markers (e.g., pronounced mtDNA structure, but little to no nuclear genetic structure) were the result of unidirectional male-mediated gene flow coupled with the absence of female-mediated gene flow between the two social forms of the fire ant S. invicta (Ross and Shoemaker, 1997; Ross et al., 1997). Similar discordance between markers and restricted female dispersal is also reported in many ant species, including, but not limited to, Cataglyphis cursor, Dorylus fulvus, and Trachymyrmex septentrionalis (Clémencet et al., 2005; Barth et al., 2013; Matthews et al., 2021). In S. daguerrei, we found four distinct genetic clusters based on nuclear markers, and one major clade of mtDNA haplotypes containing GC1, GC2, and GC3, with GC4 forming a separate clade with S. electra and S. pusilignus. Although GC1 and GC2 are significantly differentiated (G’ST=0.40-0.54), none of our genetic markers were fully diagnostic in distinguishing the two clusters from each other. These results suggest both males and females of S. daguerrei have limited dispersal and are consistent with a previous laboratory/field experiment to assess intranidal mating in S. daguerrei, where only parasite females were reported leaving the nest during mating flights; males do not disperse and die shortly after mating within their natal nest (Calcaterra et al., 2001). We also found few instances of significant isolation by distance within the parasite genetic clusters, which suggests generally limited, but not entirely restricted, dispersal and low levels of gene flow.
Our results indicate that S. daguerrei does not parasitize one social form preferentially more than the other. This is an interesting result because polygyny is a trait in free-living ant colonies suspected to facilitate the transition into a parasitic lifestyle; multiple host queens per nest and reduced colony aggression are attractive traits to parasites as they appear easier to parasitize and can disperse with hosts via budding (Rabeling et al., 2014). However, it has been suggested that once advanced stages of social parasitism have evolved, polygyny will not necessarily be preferred (Brandt et al., 2005).
Previous field observations theorized that mating flights are rare (i.e., short distances a few meters away) or absent in S. daguerrei, highly indicating that this parasite is inbred and engages in intranidal mating (Silveira-Guido et al., 1973; Hölldobler and Wilson, 1990; Calcaterra et al., 2001). We provide genetic evidence supporting these field observations, as we observed consistent departures from HWE at markers within clusters of S. daguerrei. In contrast, we found very few instances of genotype proportions deviating from those expected under HWE for the hosts, and nearly equal observed and expected heterozygosities in the host, indicating hosts have extensive dispersal capabilities and mate with individuals outside the natal colony, which is consistent with results from previous studies (Ross and Shoemaker, 1997; Ross et al., 1997; Shoemaker et al., 2006).
Previous studies on other inquilines provide mixed results on inbreeding. Moderate inbreeding was seen among Myrmica microrubra populations (Vepsäläinen et al., 2009), but it was not recorded in inquiline P. xene; however, the study was performed on a small sample size (Trontti et al., 2006). Inbreeding is also rare or nonexistent in the incipient inquiline Acromyrmex insinuator, which is not surprising as this species has not yet evolved advanced inquiline characteristics and still participates in mating flights (Sumner et al., 2004; Rabeling and Bacci, 2010).
The genome-wide departures of observed genotype proportions from H-W expectations in S. daguerrei populations are consistent with high levels of inbreeding and suggest the species may have adopted an alternative reproductive system, such as clonality, to mitigate the adverse effects associated with extensive inbreeding. Clonality, characterized by reproductive queens and males being genetic replicas of their parents, has been reported in several ant species, including Wasmannia auropunctata, Paratrechina longicornis, and Solenopsis geminata, among others (Fournier et al., 2005; Pearcy et al., 2011; Chifflet et al., 2018; Lacy et al., 2019). Whether clonality exists in S. daguerrei is unknown. Both tests performed for clonality (number of MLGs and estimates of clonal population structure) overall generally suggest S. daguerrei produces queen offspring sexually, with the possible exception of ants in GC4. If clonality was present, we would expect a pronounced drop in the number of observed multilocus genotypes in comparison to the number of individuals l (G/N ratio close to zero) and significant clonal population structure. In contrast, the ratio between the observed multilocus genotypes and the total number of individuals (G/N) among each genetic cluster and genetic cluster partitioned at individual sites ranges from 0.89-1.0 and no significant clonal structure was evident in any of the clusters with the possible exception of GC4, which is consistent with sexual reproduction in this parasite. We also evaluated this ratio among multiple winged female S. daguerrei from the same colony and, similarly, did not observe a large drop in multilocus genotypes in comparison to number of individuals (i.e., G/N ratio close to 1). Similar G/N ratios are seen in the fire ant hosts, which are known to exhibit sexual reproduction. Also, the contrasting patterns of genetic differentiation in the “Within GC, allopatric” scenario, specifically high G’ST, but little to no differentiation in the mitochondrial genome (ɸST), is more in line with expected patterns for sexually reproducing organisms, especially when one considers the effects of Wolbachia. In sexually reproducing organisms, one would predict the maternally inherited mtDNA genome would have extremely reduced variation because of hitchhiking with Wolbachia, whereas the nuclear genome would be largely unaffected. However, in a clonal organism infected with Wolbachia, we would expect complete or nearly complete linkage of the nuclear and mitochondrial genomes due to lack of recombination, and, as a result, reduced variation in both genomes. We do not see the latter pattern in our results. The potential of clonality in GC4 warrants deeper investigation, as it could be a factor contributing to the genetic distinctiveness of GC4 compared with the other S. daguerrei clusters. Also, conflicting outcomes from different analyses suggest further studies are necessary to confirm clonality in this species, which include laboratory studies that involve genotyping mated queens and their known offspring.
Our inferences based on genetic data regarding the breeding and dispersal biology of S. daguerrei match previous observations regarding the lifestyle of inquiline social parasites, in that they are highly inbred and engage in intranidal mating due to poor dispersal morphology (small body size and rudimentary wings) (Wilson, 1971; Hölldobler and Wilson, 1990; de la Mora et al., 2020). These inferences may also explain other results obtained in this study, such as host association and genetic differentiation among S. daguerrei populations. Given their poor dispersal biology and small body size, inquiline prevalence is predicted to be variable and dependent on the frequencies and distributions of their hosts. Extreme specialization on a specific host also may facilitate divergence between populations, eventually resulting in host races or entirely different species. Our results indicate highly reduced gene flow and significant differentiation among S. daguerrei populations.
5 Conclusion
Obligate inquiline social parasites are ideal for studying host-parasite co-evolution, as they parasitize closely related hosts and share the same environment with their hosts throughout their lifetime. To our knowledge, our study is the first to describe the population genetic structure of the inquiline social parasite Solenopsis daguerrei. We provide genetic evidence that S. daguerrei comprises several distinct genetic clusters, with each displaying varying degrees of host association across its known range. The significant genetic differentiation among genetic clusters and the lack of significant gene flow between them, even in sympatry, reinforce the suggestion of genetically distinct groups (presumably cryptic species) within nominal S. daguerrei. Inbreeding is common in this inquiline, which is consistent with a classic characteristic of the inquiline lifestyle. Knowledge gained from this study is crucial for informing ongoing or future biological control research, as the patterns of host association shown in our study can be used in guiding the collection of inquilines for use in a biological control program aimed at reducing populations of the highly invasive pest S. invicta.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI Genbank, Dag5: OR699212, Dag6: OR699213, Dag7: OR699214, Dag8: OR699215, Dag9: OR699216, Dag10: OR699217, Dag13: OR699218.
Author contributions
AD performed data analyses and wrote the paper. SR performed data analyses. AB aided in designing the experiment and sampling scheme, obtained collection permits, collected many of the samples, and performed data analyses. LC proposed the study model and collecting sites, and aided in most sample collections. RS aided in obtaining collection permits and in sample collection. MS aided in data analyses. DS designed the experiment and sampling scheme, aided in sample collection and data analyses, and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
The author/s declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from the US Department of Agriculture NRICGP (2006-35302-17451; 2006-35302-18001).
Acknowledgments
We would like to thank the lab members of Drs. Kenneth Ross, Brendan Hunt, and Michael Goodisman from the University of Georgia and Georgia Tech for their helpful comments on an earlier draft of this manuscript.
Conflict of interest
SR is employed by company Exponent.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1227847/full#supplementary-material
References
Ahrens M. E., Shoemaker D. (2005). Evolutionary History of Wolbachia infections in the fire ant Solenopsis invicta. BMC Evolutionary Biol. 5 (35). doi: 10.1186/1471-2148-5-35
Ahrens M. E., Ross K. G., Shoemaker D. D. (2005). Phylogeographic structure of the fire ant Solenopsis invicta in its native South American range: Roles of natural barriers and habitat connectivity. Evolution 59 (8), 1733–1743.
Aktas C. (2020). R Package haplotypes: Manipulating DNA sequences and estimating unambiguous haplotype network with statistical parsimony version 1.1.2 (CRAN).
Aron S., Passera L., Keller L. (2004). Evolution of miniaturisation in inquiline parasitic ants: Timing of male elimination in Plagiolepis pygmaea, the host of Plagiolepis xene. Insectes Sociaux 51 (4), 395–399. doi: 10.1007/s00040-004-0758-9
Ascunce M., Bouwma A. M., Shoemaker D. (2009). Characterization of 24 microsatellite markers in eleven species of fire ants in the genus Solenopsis (Hymenoptera: Formicidae). Mol. Ecol. Resour. 9 (6), 1475–1479. doi: 10.1111/j.1755-0998.2009.02688.x
Ascunce M. S., Yang C. C., Oakey J., Calcaterra L., Wu W. J., Shih C. J., et al. (2011). Global invasion history of the fire ant Solenopsis invicta. Science 331, 1066–1068. doi: 10.1126/science.1198734
Barth M. B., Moritz R. F. A., Pirk C. W. W., Kraus F. B. (2013). Male-biased dispersal promotes large scale gene flow in a subterranean army ant, Dorylus (Typhlopone) fulvus. Population Ecol. 55 (4), 523–533. doi: 10.1007/s10144-013-0383-4
Bekkevold D., Boomsma J. J. (2000). Evolutionary transition to a semelparous life history in the socially parasitic ant Acromyrmex insinuator. J. Evol. Biol. 13, 615–623. doi: 10.1046/j.1420-9101.2000.00201.x
Borgmeier T. (1949). Formigas novas ou pouco conhecidas de Costa Rica e da Argentina (Hymenoptera, Formicidae). Rev. Bras. Biol. 9, 201–210.
Borowiec M. L., Cover S. P., Rabeling C. (2021). The evolution of social parasitism in Formica ants revealed by a global phylogeny. PNAS 118 (38). doi: 10.1073/pnas.2026029118
Brandt M., Foitzik S., Fischer-Blass B., Heinze J. (2005). The coevolutionary dynamics of obligate ant social parasite systems—Between prudence and antagonism. Biol. Rev. Cambridge Philos. Soc. 80 (2), 251–267. doi: 10.1017/s1464793104006669
Briano J., Calcaterra L., Varone L. (2012). Fire ants (Solenopsis spp.) and their natural enemies in southern South America. Psyche 2012, 198084. doi: 10.1155/2012/198084
Briano J. A., Calcaterra L. A., Williams D. F., Oi D. H. (2002). Attempts to artificially propagate the fire ant parasite Solenopsis daguerrei (Hymenoptera: Formicidae) in Argentina. Florida Entomologist 85 (3), 518–520. doi: 10.1653/0015-4040(2002)085[0518:ATAPTF]2.0.CO;2
Briano J. A., Calcaterra L. A., Wojcik D. P., Williams D. F., Banks W. A. (1997). Abundance of the parasitic ant Solenopsis daguerrei (Hymenoptera: Formicidae) in South America, a potential candidate for the biological control of the red imported fire ant in the United States. Environ. Entomology 26 (5), 1143–1148. doi: 10.1093/ee/26.5.1143
Bromham L., Leys R. (2005). Sociality and the rate of molecular evolution. Mol. Biol. Evol. 22 (6), 1393–1402. doi: 10.1093/molbev/msi133
Bruch C. (1930). Notas preliminares acerca de Labauchena daguerrei Santschi. Rev. Soc Entomol. Argent. 3, 73–82.
Brunton A., Lee D. J., Conroy G., Ogbourne S. (2021). Clonal reproduction and low genetic diversity in northern Australian Santalum lanceolatum (Santalaceae) populations highlights the need for genetic rescue of this commercially significant species. Forests 12 (6), 741. doi: 10.3390/f12060741
Burgess S. L., Gilchrist C. A., Lynn T. C., Petri J. W. A. (2017). Parasitic protozoa and interactions with the host intestinal microbiota. Infect. Immun. 85 (8). doi: 10.1128/IAI.00101-17
Buschinger A. (1986). Evolution of social parasitism in ants. Trends Ecol. Evol. 1 (6), 155–160. doi: 10.1073/pnas.2026029118
Buschinger A. (1990). Sympatric speciation and radiative evo-lution of socially parasitic ants – heretic hypotheses and their factual background. Z. für Zoologische Systematik und Evolutionsforschung. 24, 241–260.
Buschinger A. (2009). Social parasitism among ants: A review (Hymenoptera: Formicidae). Myrmecological News 12, 219–235.
Calcaterra L. A., Briano J. A., Williams D. F. (1999). Field studies of the parasitic ant Solenopsis daguerrei (Hymenoptera: Formicidae) on fire ants in Argentina. Environ. Entomology 28 (1), 88–95. doi: 10.1093/ee/28.1.88
Calcaterra L. A., Briano J. A., Williams D. F. (2000). New host for the parasitic ant Solenopsis daguerrei (Hymenoptera: Formicidae) in Argentina. Florida Entomologist 83 (3), 363–365. doi: 10.2307/3496358
Calcaterra L. A., Briano J. A., Williams D. F., Oi D. H. (2001). Observations on the sexual castes of the fire ant parasite Solenopsis daguerrei (Hymenoptera: Formicidae). Florida Entomologist 84 (3), 446–448. doi: 10.2307/3496509
Chen Y. P., Lu L. Y., Skow L. C., Vinson S. B. (2003). Relatedness among co-existing queens within polygyne colonies of a Texas population of the fire ant, Solenopsis invicta. Southwestern Entomol 28, 27–36.
Chifflet L., Guzman N. V., Rey O., Confalonieri V. A., Calcaterra L. A. (2018). Southern expansion of the invasive ant Wasmannia auropunctata within its native range and its relation with clonality and human activity. PLoS One 13 (11). doi: 10.1371/journal.pone.0206602
Cini A., Sumner S., Cervo R. (2019). Inquiline social parasites as tools to unlock the secrets of insect sociality. Phil. Trans. R. Soc 374. doi: 10.1098/rstb.2018.0193
Clémencet J., Viginier B., Doums C. (2005). Hierarchical analysis of population genetic structure in the monogynous ant Cataglyphis cursor using microsatellite and mitochondrial DNA markers. Mol. Ecol. 14 (12), 3735–3744. doi: 10.1111/j.1365-294X.2005.02706.x
Dedeine F., Ahrens M., Calcaterra L., Shoemaker D. D. (2005). Social parasitism in fire ants (Solenopsis spp.): A potential mechanism for interspecies transfer of Wolbachia. Mol. Ecol. 14 (5), 1543–1548. doi: 10.1111/j.1365-294X.2005.02499.x
Degueldre F., Mardulyn P., Kuhn A., Pinel A., Karaman C., Lebas C., et al. (2021). Evolutionary history of inquiline social parasitism in Plagiolepis ants. Mol. Phylogenet. Evol. 155, 107016. doi: 10.1016/j.ympev.2020.107016
de la Mora A., Sankovitz M., Purcell J. (2020). Ants (Hymenoptera: Formicidae) as host and intruder: Recent advances and future directions in the study of exploitative strategies. Myrmecol. News 30, 53–71. doi: 10.25849/myrmecol.news_030:053
Deng J., Assandri G., Chauhan P., Futahashi R., Galimberti A., Hansson B., et al. (2021). Wolbachia-driven selective sweep in a range expanding insect species. BMC Ecol. Evol. 21 (181), 1–17. doi: 10.1186/s12862-021-01906-6
Durden L. A., Musser G. G. (1994). The sucking lice (Insecta: Anoplura) of the world: A taxonomic checklist with records of mammalian hosts and geographical distributions. Bull. American Museum of Natural History 218.
Earl D. A., vonHoldt B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4 (2), 359–361. doi: 10.1007/s12686-011-9548-7
Emery C. (1909). Über den Ursprung der dulotischen, parasitischen una myrmekophilen Ameisen. Biologisches Centralblatt 29, 352–362.
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Fournier D., Estoup A., Orivel J., Foucaud J., Jourdan H., Breton J. L., et al. (2005). Clonal reproduction by males and females in the little fire ant. Nature 435 (7046), 1230–1234. doi: 10.1038/nature03705
Franks N., Blum M., Smith R. K., Allies A. B. (1990). Behavior and chemical disguise of cuckoo ant Leptothorax kutteri in relation to its host Leptothorax acervorum. J. Chem. Ecol. 16 (5), 1431–1444. doi: 10.1007/BF01014079
Gallardo A. (1916). Notes systematiques et ethologiques sur les fourmis attines de la Republique Argentine. Anales del Museo Nacional Hist. Natural Buenos Aires 28, 317–344.
Glatzel G., Geils B. W. (2009). Mistletoe ecophysiology: Host-parasite interactions. Botany 87, 10–15. doi: 10.1139/B08-096
Gotzek D., Axen H. J., Suarez A. V., Helms Cahan S., Shoemaker D. (2015). Global invasion history of the tropical fire ant: A stowaway on the first global trade routes. Mol. Ecol. 24 (2), 374–388. doi: 10.1111/mec.13040
Goudet J. (2005). Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5 (1), 184–186. doi: 10.1111/j.1471-8286.2004.00828.x
Hedrick P. W. (2005). A standardized genetic differentiation measure. Evolution 59 (8), 1633–1638. doi: 10.1111/j.0014-3820.2005.tb01814.x
Huelsenbeck J. P., Ronquist F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinf. (Oxford England) 17 (8), 754–755. doi: 10.1093/bioinformatics/17.8.754
Jansen G., Savolainen R., Vepsäläinen K. (2010). Phylogeny, divergence-time estimation, biogeography and social parasite-host relationships of the Holarctic ant genus Myrmica (Hymenoptera: Formicidae). Mol. Phylogenet. Evol. 56 (1), 294–304. doi: 10.1016/j.ympev.2010.01.029
Jiggins F. M. (2003). Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics 164, 5–12. doi: 10.1093/genetics/164.1.5
Jombart T., Devillard S., Balloux F. (2010). Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 11 (94), 1–15. doi: 10.1186/1471-2156-11-94
Jombart T., Devillard S., Dufour A. B., Pontier D. (2008). Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101 (1), 92–103. doi: 10.1038/hdy.2008.34
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Kamvar Z. N., Tabima J. F., Grünwald N. J. (2014). Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281. doi: 10.7717/peerj.281
Keller L., Passera L. (1989). Size and fat content of gynes in relation to the mode of colony founding in ants (Hymenoptera; Formicidae). Oecologia 80 (2), 236–240. doi: 10.1007/BF00380157
Kopelman N. M., Mayzel J., Jakobsson M., Rosenberg N. A., Mayrose I. (2015). Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15 (5), 1179–1191. doi: 10.1111/1755-0998.12387
Krieger M. J. B., Keller L. (1997). Polymorphism at dinucleotide microsatellite loci in fire ant Solenopsis invicta populations. Mol. Ecol. 6 (10), 997–999. doi: 10.1046/j.1365-294X.1997.00264.x
Krieger M. J. B., Ross K. G. (2002). Identification of a major gene regulating complex social behavior. Science 295 (5553), 328–332. doi: 10.1126/science.1065247
Lacy K. D., Shoemaker D., Ross K. G. (2019). Joint evolution of asexuality and queen number in an ant. Curr. Biol. 29, 1394–1400. doi: 10.1016/j.cub.2019.03.018
Lafferty K. D. (2008). “Parasites,” in Encyclopedia of Ecology. Eds. Jørgensen S. E., Fath B. D. (Elsevier Science: Academic Press), 2640–2644.
Lenoir A., D’Ettorre P., Errard C. (2001). Chemical ecology and social parasitism in ants. Annu. Rev. Entomol. 46, 573–599. doi: 10.1146/annurev.ento.46.1.573
Leppänen J., Seppä P., Vepsäläinen K., Savolainen R. (2016). Mating isolation between the ant Myrmica rubra and its microgynous social parasite. Insectes Sociaux 63 (1), 79–86. doi: 10.1007/s00040-015-0438-y
Maddison W. P., Maddison D. R. (2019). Mesquite: a modular system for evolutionary analysis. Version 3.6.1. Available at: http://mesquiteproject.org.
Mardulyn P., Thurin N., Piou V., Grumiau L., Aron S. (2014). Dispersal in the inquiline social parasite ant Plagiolepis xene. Insect. Soc 61, 197–202. doi: 10.1007/s00040-014-0345-7
Matthews A. E., Kellner K., Seal J. N. (2021). Male-biased dispersal in a fungus-gardening ant symbiosis. Ecol. Evol. 11 (5), 2307–2320. doi: 10.1002/ece3.7198
Matute D. R., Cooper B. S. (2021). Comparative studies on speciation: 30 years since Coyne and Orr. Evolution 75 (4), 764–778. doi: 10.1111/evo.14181
Meirmans P. G. (2015). Seven common mistakes in population genetics and how to avoid them. Mol. Ecol. 24 (13), 3223–3231. doi: 10.1111/mec.13243
Meirmans P. G. (2020). Genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20 (4), 1126–1131. doi: 10.1111/1755-0998.13145
Meirmans P. G., van Tienderen P. H. (2004). genotype and genodive: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4 (4), 792–794. doi: 10.1111/j.1471-8286.2004.00770.x
Mera-Rodríguez D., Jourdan H., Ward P., Shattuck S., Cover S., Wilson E., et al. (2023). Biogeography and evolution of social parasitism in Australian Myrmecia bulldog ants revealed by phylogenomics. Mol. Phylogenet. Evol. 186, 107825. doi: 10.1016/j.ympev.2023.107825
Miller M. A., Pfeiffer W., Schwartz T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 14 Nov. 2010. 1–8. Available at: https://www.phylo.org/sub_sections/portal/cite.php.
Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Nylander J. A. A. (2004). MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre (Uppsala University). Available at: https://github.com/nylander/MrModeltest2.
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2020). Vegan community ecology package version 2.5-7 (CRAN).
O’Neill S. L., Giordano R., Colbert A. M. E., Karr T. L., Robertson H. M. (1992). 16S rRNA Phylogenetic Analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. United States America 89 (7), 2699–2702. doi: 10.1073/pnas.89.7.2699
Paradis E. (2010). pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 26 (3), 419–420. doi: 10.1093/bioinformatics/btp696
Passera L., Gilbert M., Aron S. (2001). Social parasitism in ants: Effects of the inquiline parasite Plagiolepis xene St. on queen distribution and worker production of its host Plagiolepis pygmaea Latr. Insects Soc 48, 74–79. doi: 10.1007/PL00001749
Pearcy M., Goodisman M. A. D., Keller L. (2011). Sib mating without inbreeding in the longhorn crazy ant. Proc. R. Soc. B 278, 2677–2681. doi: 10.1098/rspb.2010.2562
Pesquero M. A., Penteado-Dias A. M. (2004). New records of Orasema xanthopus (Hymenoptera: Eucharitidae) and Solenopsis daguerrei (Hymenoptera: Formicidae) from Brazil. Braz. J. Biol. 64 (3B), 737.
Petrie M., Møller A. P. (1991). Laying eggs in others’ nests: intraspecific brood parasitism in birds. Trends Ecol. Evol. 6 (10), 315–320. doi: 10.1016/0169-5347(91)90038-Y
Pitts J. P. (2002). A cladistic analysis of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). PhD thesis (USA: University of Georgia).
Pitts J. P., Camacho G. P., Gotzek D., McHugh J. V., Ross K. G. (2018). Revision of the fire ants of the Solenopsis saevissima species group (Hymenoptera: Formicidae). Proc. Entomological Soc. Washington 120, 308–411. doi: 10.4289/0013-8797.120.2.308
Pitts J. P., McHugh J. V., Ross K. G. (2005). Cladistic analysis of the fire ants of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). Zoologica Scripta 34 (5), 493–505. doi: 10.1111/j.1463-6409.2005.00203.x
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Rabeling C., Bacci J. M. (2010). A new workerless inquiline in the Lower Attini (Hymenoptera: Formicidae), with a discussion of social parasitism in fungus-growing ants. Systematic Entomology 35, 379–392. doi: 10.1111/j.1365-3113.2010.00533.x
Rabeling C., Schultz T. R., Pierce N. E., Bacci M. Jr. (2014). A social parasite evolved reproductive isolation from its fungus-growing ant host in sympatry. Curr. Biol. 24, 2047–2052. doi: 10.1016/j.cub.2014.07.048
Ross K. G. (1997). Multilocus evolution in fire ants: Effects of selection, gene flow and recombination. Genetics 145 (4), 961–974. doi: 10.1093/genetics/145.4.961
Ross K. G., Keller L. (1998). Genetic control of social organization in an ant. Proc. Natl. Acad. Sci. 95 (24), 14232–14237. doi: 10.1073/pnas.95.24.14232
Ross K. G., Krieger M. J. B., Shoemaker D. D. (2003). Alternative genetic foundations for a key social polymorphism in fire ants. Genetics 165 (4), 1853–1867. doi: 10.1093/genetics/165.4.1853
Ross K. G., Krieger M. J. B., Shoemaker D. D., Vargo E. L., Keller L. (1997). Hierarchical analysis of genetic structure in native fire ant populations: results from three classes of molecular markers. Genetics 147, 643–655. doi: 10.1093/genetics/147.2.643
Ross K. G., Shoemaker D. D. (1997). Nuclear and mitochondrial genetic structure in two social forms of the fire ant Solenopsis invicta: insights into transitions to an alternate social organization. Heredity 78, 590–602. doi: 10.1038/hdy.1997.98
Ross K., Shoemaker D. D. (2005). Species delimitation in native South American fire ants. Mol. Ecol. 14, 3419–3438. doi: 10.1111/j.1365-294X.2005.02661.x
Ross K. G., Trager J. C. (1990). Systematics and population genetics of fire ants (Solenopsis saevissima complex) from Argentina. Evolution 44 (8), 2113–2134. doi: 10.1111/j.1558-5646.1990.tb04316.x
Santschi F. (1930). Un nouveau genre de fourmi parasite sans ouvriéres de l’Argentine. Rev. Soc Entomol. Argent. 3, 81–83.
Sapp J. R., Yost J., Lyon B. E. (2020). The socially parasitic ant Polyergus mexicanus has host-associated genetic population structure and related neighboring colonies. Mol. Ecol. 29, 2050–2062. doi: 10.1111/mec.15468
Schultz T. R., Bekkevold D., Boomsma J. J. (1998). Acromyrmex insinuator new species: an incipient social parasite of fungus growing ants. Insectes Sociaux 45, 457–471. doi: 10.1007/s000400050101
Shoemaker D. (2016). Development of genetic markers distinguishing two invasive fire ant species (Hymenoptera: Formicidae) and their hybrids. Florida Entomologist 99 (1), 117–119. doi: 10.1653/024.099.0122
Shoemaker D. D., Ahrens M. E., Ross K. G. (2006). Molecular phylogeny of fire ants of the Solenopsis saevissima species-group based on mtDNA sequences. Mol. Phylogenet. Evol. 38, 200–215. doi: 10.1016/j.ympev.2005.07.014
Shoemaker D., Ascunce M. S. (2010). A new method for distinguishing colony social forms of the fire ant, Solenopsis invicta. J. Insect Sci. 10, 73. doi: 10.1673/031.010.7301
Shoemaker D. D., Ross K. G., Keller L., Vargo E. L., Werren J. H. (2000). Wolbachia infections in native and introduced populations of fire ants (Solenopsis spp.). Insect Mol. Biol. 9 (6), 661–673. doi: 10.1046/j.1365-2583.2000.00233.x
Silveira-Guido A., Carbonell J., Crisci C. (1973). Animals associated with the Solenopsis (fire ants) complex with special reference to Labauchena daguerrei. Proc. Tall Timbers Conf. Ecol. Anim. Control Habitat Manage 4, 41–52.
Simon C., Frati F., Beckenbach A., Crespi B., Liu H., Flook P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc Am. 81, 651–701.
Singer T. L. (1998). Roles of hydrocarbons in the recognition systems of insects. Am. Zool. 38, 394–405. doi: 10.1093/icb/38.2.394
Sumner S., Aanen D. K., Delabie J., Boomsma J. J. (2004). The evolution of social parasitism in Acromyrmex leaf-cutting ants: A test of Emery’s Rule. Insect Soc 51, 37–42. doi: 10.1007/s00040-003-0723-z
Toews D. P., Brelsford A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x
Trontti K., Aron S., Sundström L. (2005). Inbreeding and kinship in the ant Plagiolepis pygmaea. Molec. Ecol. 14, 2007–2015. doi: 10.1111/j.1365-294X.2005.02529.x
Trontti K., Aron S., Sunström L. (2006). The genetic population structure of the ant Plagiolepis xene - implication for genetic vulnerability of obligate social parasites. Conserv. Genet. 7, 241–250. doi: 10.1007/s10592-005-9003-y
Tseng S. P., Wetterer J. K., Suarez A. V., Lee C. Y., Yoshimura T., Shoemaker D., et al. (2019). Genetic diversity and Wolbachia infection patterns in a globally distributed invasive ant. Front. Genet. 10 (838). doi: 10.3389/fgene.2019.00838
Tsutsui N. D. (2004). Scents of self: The expression component of self/non-self recognition systems. Annales Zoologici Fennici 41, 713–727.
Turelli M. (1994). Evolution of incompatibility-inducing microbes and their hosts. Evolution 48 (5), 1500–1513. doi: 10.1111/j.1558-5646.1994.tb02192.x
Turelli M., Hoffmann A. A. (1991). Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353 (6343), 440–442. doi: 10.1038/353440a0
Vepsäläinen K., Ebsen J. R., Savolainen R., Boomsma J. J. (2009). Genetic differentiation between the ant Myrmica rubra and its microgynous social parasite. Insectes Sociaux 56 (4), 425–437. doi: 10.1007/s00040-009-0042-0
Wang J., Wurm Y., Nipitwattanaphon M., Riba-Grognuz O., Huang Y. C., Shoemaker D., et al. (2013). A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493 (7434), 664–668. doi: 10.1038/nature11832
Werren J. H. (1997). Biology of Wolbachia. Annu. Rev. Entomology 42 (1), 587–609. doi: 10.1146/annurev.ento.42.1.587
Werren J. H., Baldo L., Clark M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6 (10), 741–751. doi: 10.1038/nrmicro1969
Williams D. F., Oi D. H., Sanford D. P., Pereira R. M., Briano J. A. (2003). Biological control of imported fire ants (Hymenoptera: Formicidae). Am. Entomologist. 49 (3), 150–163. doi: 10.1093/ae/49.3.150
Keywords: fire ants, population structure, social parasitism, inquiline, microsatellites, mitochondrial DNA
Citation: Dekovich A, Ryan S, Bouwma A, Calcaterra L, Silvestre R, Staton M and Shoemaker D (2023) Population genetic analyses reveal host association and genetically distinct populations of social parasite Solenopsis daguerrei (Hymenoptera: Formicidae). Front. Ecol. Evol. 11:1227847. doi: 10.3389/fevo.2023.1227847
Received: 23 May 2023; Accepted: 12 October 2023;
Published: 26 October 2023.
Edited by:
Alison G. Nazareno, Federal University of Minas Gerais, BrazilCopyright © 2023 Dekovich, Ryan, Bouwma, Calcaterra, Silvestre, Staton and Shoemaker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: DeWayne Shoemaker, dshoema2@utk.edu
 Allyson Dekovich
Allyson Dekovich Sean Ryan
Sean Ryan Andrew Bouwma3
Andrew Bouwma3  Luis Calcaterra
Luis Calcaterra Margaret Staton
Margaret Staton DeWayne Shoemaker
DeWayne Shoemaker