Proteomic Analysis of the Parasitic Cnidarian Ceratonova shasta (Cnidaria: Myxozoa) Reveals Diverse Roles of Actin in Motility and Spore Formation
- 1Marine Biology Department, The Leon H. Charney School of Marine Sciences, University of Haifa, Haifa, Israel
- 2Bioinformatic Unit, University of Haifa, Haifa, Israel
- 3Department of Microbiology, Oregon State University, Corvallis, OR, United States
- 4Biology Centre of the Czech Academy of Sciences, Institute of Parasitology, České Budějovice, Czechia
Myxozoans are widely distributed aquatic obligate endoparasites that were recently recognized as belonging within the phylum Cnidaria. They have complex life cycles with waterborne transmission stages: resistant, infectious spores that are unique to myxozoans. However, little is known about the processes that give rise to these transmission stages. To understand the molecular underpinnings of spore formation, we conducted proteomics on Ceratonova shasta, a highly pathogenic myxozoan that causes severe mortalities in wild and hatchery-reared salmonid fishes. We compared proteomic profiles between developmental stages from inside the fish host, and the mature myxospore, which is released into the water where it drifts passively, ready to infect the next host. We found that C. shasta contains 2,123 proteins; representing the first proteomic catalog of a myxozoan myxospore. Analysis of proteins differentially expressed between developing and mature spore stages uncovered processes that are active during spore formation. Our data highlight dynamic changes in the actin cytoskeleton, which provides myxozoan developmental stages with mobility through lamellipodia and filopodia, whereas in the mature myxospore the actin network supports F-actin stabilization that reinforces the transmission stage. These findings provide molecular insight into the myxozoan life cycle stages and, particularly, into the process of sporogenesis.
Introduction
Myxozoa is a large and widespread aquatic group of microscopic, obligate endoparasites that can cause severe ecological and economic effects (Wahli et al., 2002; Sterud et al., 2007; Hallett and Bartholomew, 2011). They have recently been recognized as belonging within the Cnidaria (reviewed in Atkinson et al., 2018a) and with more than 2000 species, they comprise about 20% of this ancient phylum (Lom and Dyková, 2006; Okamura et al., 2015; Hartikainen et al., 2016; Atkinson et al., 2018a). Myxozoans (myxosporeans) are unique within the Cnidaria, in that they are spore-forming parasites whose life cycles alternate between vertebrate (mostly fish) and invertebrate (mostly annelid) hosts. They are among the most common fish parasites (Mackenzie and Kalavati, 2014); however, they can also infect other vertebrate groups such as amphibians, reptiles, birds, and even terrestrial mammals (Hallett et al., 2015; Hartigan et al., 2016b, and references therein). Transmission occurs via two types of waterborne spores: myxospores, which are produced in the fish host and infect the invertebrate host, and actinospores, which operate in the opposite direction (Wolf and Markiw, 1984; Bartholomew et al., 1997; Canning and Okamura, 2003; Lom and Dyková, 2006).
Myxozoan parasites demonstrate extreme reduction in size and tissue complexity compared with most free-living cnidarians. Nevertheless, they retain the characteristic cnidarian stinging organelle, the nematocyst (or polar capsule), which initiates the infection process (Ben-David et al., 2016; Piriatinskiy et al., 2017). Nematocyst proteomic data is currently available for four myxozoan species (Piriatinskiy et al., 2017; Guo et al., 2020). After nematocyst discharge and attachment to the host, the motile parasite sporoplasm penetrates the epithelium and typically enters the bloodstream, where it begins proliferating. The parasite stages travel to a specific target tissue, where they differentiate and undergo sporogenesis. The sporogenic cells generate thousands of mature myxospores, which will eventually be released into the water column to infect the next host. The mature spore consists of only three cells: nematocyte, sporoplasm and spore valve. Spore formation represents a novel development of the myxozoans within the Cnidaria, and is counter to the general pattern of body plan simplification and gene loss associated with evolution of a parasite life style. Sporogenesis has been studied extensively using microscopy (reviewed in Feist et al., 2015); however, there are limited molecular data on this process (Alama-Bermejo et al., 2012, 2019, 2020) and the proteins that make the mature myxospores have not yet been studied.
The myxozoan Ceratonova shasta, an endemic species in the Pacific Northwest of the United States, is one of the most virulent myxozoans, causing high mortalities in wild and hatchery-reared populations of salmon and trout (Hallett et al., 2012). The parasite invades the fish gills and migrates via the blood to the intestine, where sporogenesis takes place (Bjork and Bartholomew, 2010). Parasite development causes intestinal inflammation and hemorrhage, leading to enteronecrosis, a lethal disease (Figure 1A). C. shasta is one of a few myxozoan species for which both vertebrate and invertebrate hosts are known, and can be maintained in the laboratory (Bartholomew et al., 1997), thus enabling controlled dosing and infection experiments. Here, we used this parasite as a model organism to discover molecular processes that occur during sporogenesis. Taking a proteomic approach, we isolated developmental stages from the fish ascites and compared them to mature myxospores. Our study provides the first catalog of myxozoan proteins and highlights some of the active pathways intrinsic to creation of mature myxospores.
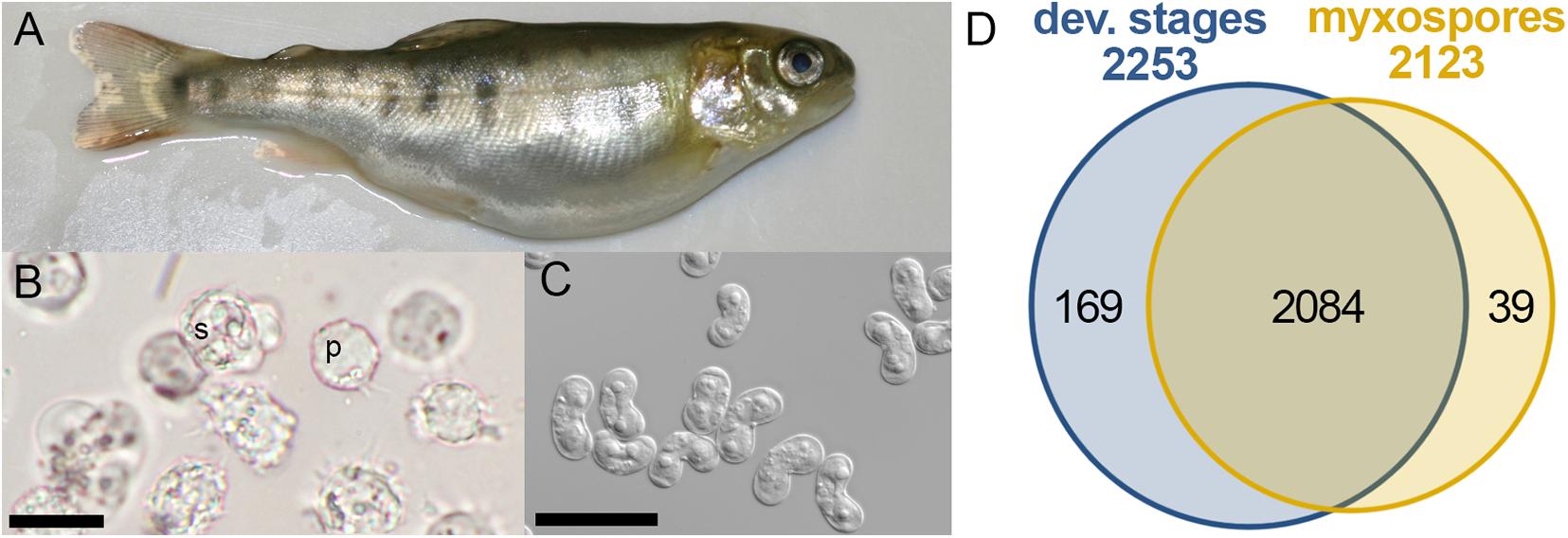
Figure 1. Comparative proteomics of Ceratonova shasta stages from fish. (A) Infected fish showing bloated abdomen typical of ascites production. (B) Developmental stages extracted from ascites: pre-sporogonic, p; sporogonic, s; bar 20 μm. (C) Purified mature myxospores; bar 20 μm. (D) Venn diagram of identified protein groups in developmental stages and mature myxospores.
Materials and Methods
Isolation and Extraction of Developmental Stages and Mature Myxospores
C. shasta has different host-associated genotypes that differ in virulence (Atkinson and Bartholomew, 2010; Stinson et al., 2018; Alama-Bermejo et al., 2020). Genotype IIR infections are highly virulent and become systemic, inducing the production of large volumes of ascites in the body cavity of susceptible rainbow trout. This fluid is rich in different developmental stages of C. shasta (pre-sporogonic and sporogonic) and an easy source of a large amount of parasites. In this study, we used genotype IIR C. shasta from rainbow trout that had been infected by intraperitoneal injection of parasites from a donor fish (Atkinson et al., 2018b). Ascites (2–7 mL/fish) was collected by syringe from the abdominal cavity of 5 infected fish, then individually centrifuged, and the pellets suspended in PBS and centrifuged again. The supernatant was discarded and the sample pellets frozen and lyophilized. From 3 fish, mature myxospores were isolated from intestine samples. Because of limited material, only three biological samples of myxospores were purified using a Percoll gradient as described (Piriatinskiy et al., 2017); each sample contained about 2 million myxospores.
Proteolysis of developmental stages, and mature myxospores was performed for 1 h at room temperature (RT) with 1% SDS. Then, 0.1 M DTT was added for 30 min at 60°C and the samples were modified with 0.4 M iodoacetamide for 30 min in the dark at RT and precipitated with 80% acetone. The resultant pellet was suspended in buffer (9 M urea, 0.4 M ammonium bicarbonate) and digested in 1 M urea, 0.06 mM ammonium bicarbonate with modified trypsin (Promega) at a 1:50 enzyme-to-substrate ratio overnight at 37°C.
All experiments performed using live animals were in accordance with state and federal regulations, and were pre-approved by the Oregon State University’s Institutional Animal Care and Use Committee (IACUC; protocol #5040).
Mass Spectrometry Analysis
Eight individual samples were analyzed: five ascites samples and three myxospore samples. The tryptic peptides were desalted using C18 tips (Toptip, Glygen), dried and resuspended in 0.1% formic acid. Two micrograms of the resultant peptides were resolved by reverse-phase chromatography on 0.075 X 180-mm fused silica capillaries (J&W) packed with Reprosil reversed phase material (Dr. Maisch GmbH, Germany). The peptides were eluted with linear 120 min gradient of 5–28%, 15 min gradient of 28–95% and 15 min at 95% acetonitrile with 0.1% formic acid in water at a flow rate of 0.15 μl/min. Mass spectrometry was performed with Q Exactive plus mass spectrometer (Thermo) in a positive mode (m/z 300–1,800, resolution 70,000) using repetitive full MS scan followed by collision induced dissociation (HCD, at 25 eV normalized collision energy, isolation window 1.8 M/Z) of the 10 dominant ions (>1 charges) selected from the first MS scan. A dynamic exclusion list was enabled with exclusion duration of 20 s.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD022770 and 10.6019/PXD022770.
Sporogenesis and Mature Spore Analysis
The mass spectrometry data from the biological replicates were label-free analyzed and quantified using MaxQuant (ver. 1.5.2.8) for peak picking identification and quantitation using the Andromeda search engine, searching against the database with mass tolerance of 20 and 4.5 ppm after calibration. Oxidation on methionine, and protein N-terminus acetylation were accepted as variable modifications and carbamidomethyl on cysteine was accepted as static modifications. Minimal peptide length was set to seven amino acids and a maximum of two miscleavages was allowed. Peptide- and protein-level false discovery rates (FDRs) were filtered to 1% using the target-decoy strategy. Protein table were filtered to eliminate identifications from the reverse database, and common contaminants. The databases that were used were the Oncorhynchus mykiss proteins from Uniprot (accessed December 20, 2017) and the Open Reading Frames (ORFs) of the C. shasta transcriptome “IIR RBT6 cs neither assembly” (Alama-Bermejo et al., 2020), with 1% FDR. The ORFs were generated using TransDecoder1, set at a minimum length of 30 amino acids. C. shasta identified ORFs were clustered using CD-hit (Fu et al., 2012), to remove redundant identical ORFs or segments of larger ORFs. When several ORFs were detected in a single transcript, only ORFs with length ≥ 50% of the longest detected ORF were selected. C. shasta proteins annotation and interpro domains were obtained using Blast2GO (ver. 5.1.12) (default settings—02.2019) and using blastp, protein homologs of Mus musculus were identified for STRING analysis. BUSCO (version 4.1.4) was used to evaluate the comprehensiveness of the proteomic dataset (Simão et al., 2015).
Downstream proteomic analyses were performed using Perseus (ver. 1.5.0.31). Protein groups were filtered to eliminate the identifications from the reverse database, common contaminants and single peptide identifications. Additionally, proteins identified as fish proteins were filtered out. The protein identification cutoff was set to at least two spectra containing one unique peptide. Hits in only one biological sample were dismissed. Normalized LFQ intensities were log2-transformed and missing values were imputed with constant value of 18. The ANOVA multiple sample test using a permutation-based FDR of 0.05 was performed to identify statistically significant differences between the two groups. A heat map showing z-scored log-transformed LFQ intensities of differentially expressed proteins was created using R package “superheat” (ver. 0.1.0). Interactions of proteins from developmental stages and myxospores were examined by STRING (ver. 11) (Jensen et al., 2009), which is capable of inferring protein-protein interactions from homologs of mouse proteins, using default parameters of medium confidence (0.4). STRING was also used for GO analysis of molecular function enrichment using mouse as the background.
Results
Proteomic Analysis
To identify changes in the proteome between developing parasites (pre-sporogonic and sporogonic stages) and the mature myxospore, we carried out a comparative proteomic profiling. We analyzed proteomes of 5 replicates of developmental stages, and three of isolated mature myxospores (Figures 1A–C). Our analysis focused on proteins that were identified with high confidence in at least two of the samples of the different parasite stages. We identified 2,292 protein groups of which we could annotate 2,163 (Supplementary Tables 1, 2). From these, 2,084 proteins (91%) were common to both developmental stages and mature spores, 169 (7.4%) were specific to developmental stages, and 39 (1.7%) were specific to mature spores (Figure 1D). Supplementary Table 1 lists these proteins, their abundance and annotations. The completeness of the proteomic analysis was assessed using the BUSCO subset of 255 widely conserved eukaryotic core proteins (Simão et al., 2015). The analysis indicated that 35.3% of the core proteins were identified and an additional 6.3% were fragmented resulting in a total of 41.6% representation of the core protein in the proteome.
The Most Abundant Proteins During Sporogenesis Are Related to Metabolism
Comparison of the 100 most highly expressed proteins from developmental stages and mature myxospores showed that metabolism-related and ribosomal proteins were more highly expressed during sporogenesis (Figure 2). Proteins related to the cytoskeleton were also more abundant in developmental stages than in the mature myxospores. Interestingly, unknown proteins and nematocyst proteins were more abundant in the myxospores, although nearly all identified nematocyst-related proteins were found in both fractions. These findings are the expected activity of the parasite as it builds spores.
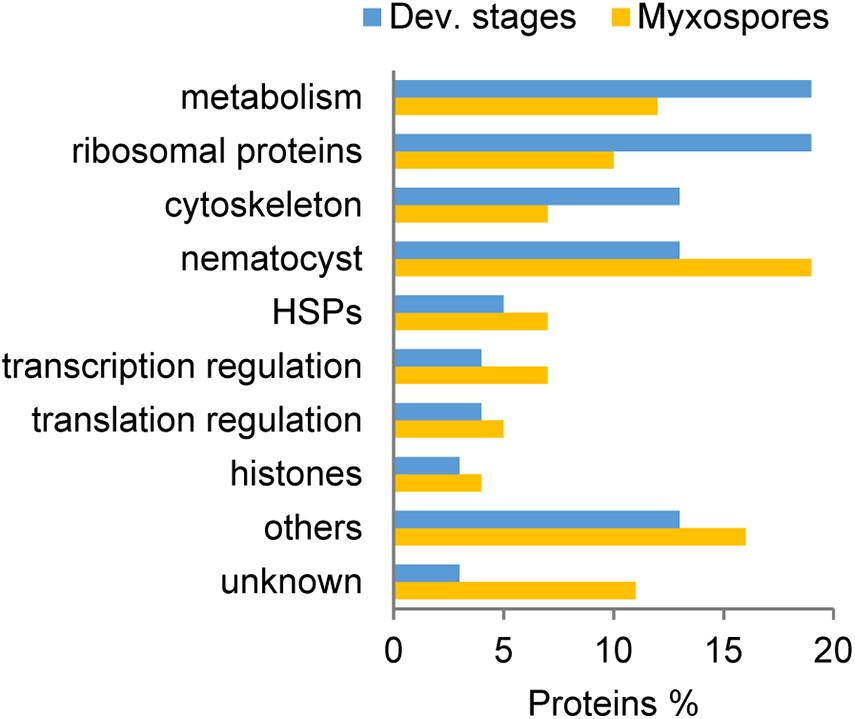
Figure 2. Comparison of the 100 most abundant protein groups in developmental stages (Dev. stages) and mature myxospores. Proteins were grouped based on putative functions. HSPs: heat-shock proteins, unknown- with no annotation.
Specific Protein Expression During Sporogenesis and in Mature Myxospores
To compare protein expression profiles between the 2,084 proteins found in both developmental stages and mature spores, we performed ANOVA tests. We found 357 protein groups with statistically significant stage-specific expression (FDR < 0.05) (Figure 3 and Supplementary Table 3). To find the net interaction and the pathways to which the stage-specific expressed proteins are related, we searched the STRING database for C. shasta mouse homologs. We combined the stage-specific proteins (Figure 1D) with the common significantly highly expressed proteins (Figure 3) for each stage and analyzed the data for GO molecular function enriched terms (FDR adjusted P < 0.05). The analysis revealed 25 enriched GO terms specific to the developmental stages, which were related to actin and cytoskeleton, GTPase, endopeptidase activity, ribonucleotide binding and other processes (Figure 4A). In the mature spores, we found five specifically enriched GO terms, including protein kinase binding, ion binding and RNA binding (Figure 4B). We identified 17 enriched GO terms shared by both sample types, and these were related mainly to binding and catalytic activity (Figure 4C).
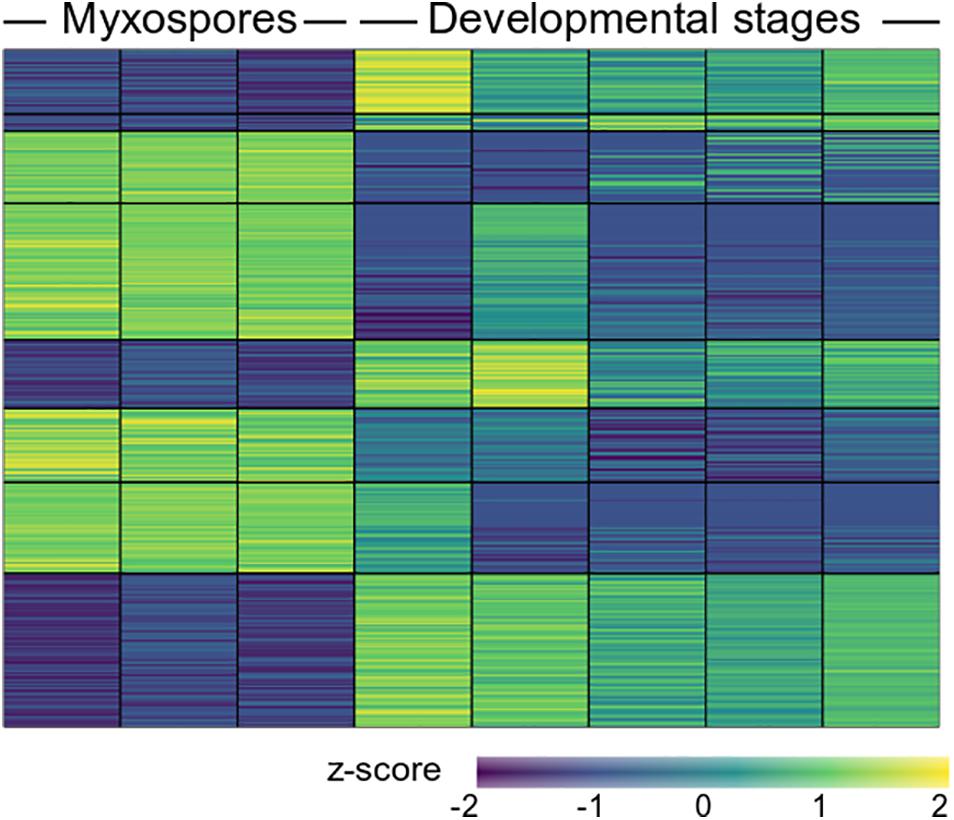
Figure 3. Heat map showing 357 differentially expressed proteins (FDR adjusted P < 0.05) between mature myxospores and developmental stages (n = 3 and n = 5 biological replicates, respectively). The full protein details are in Supplementary Table 3.
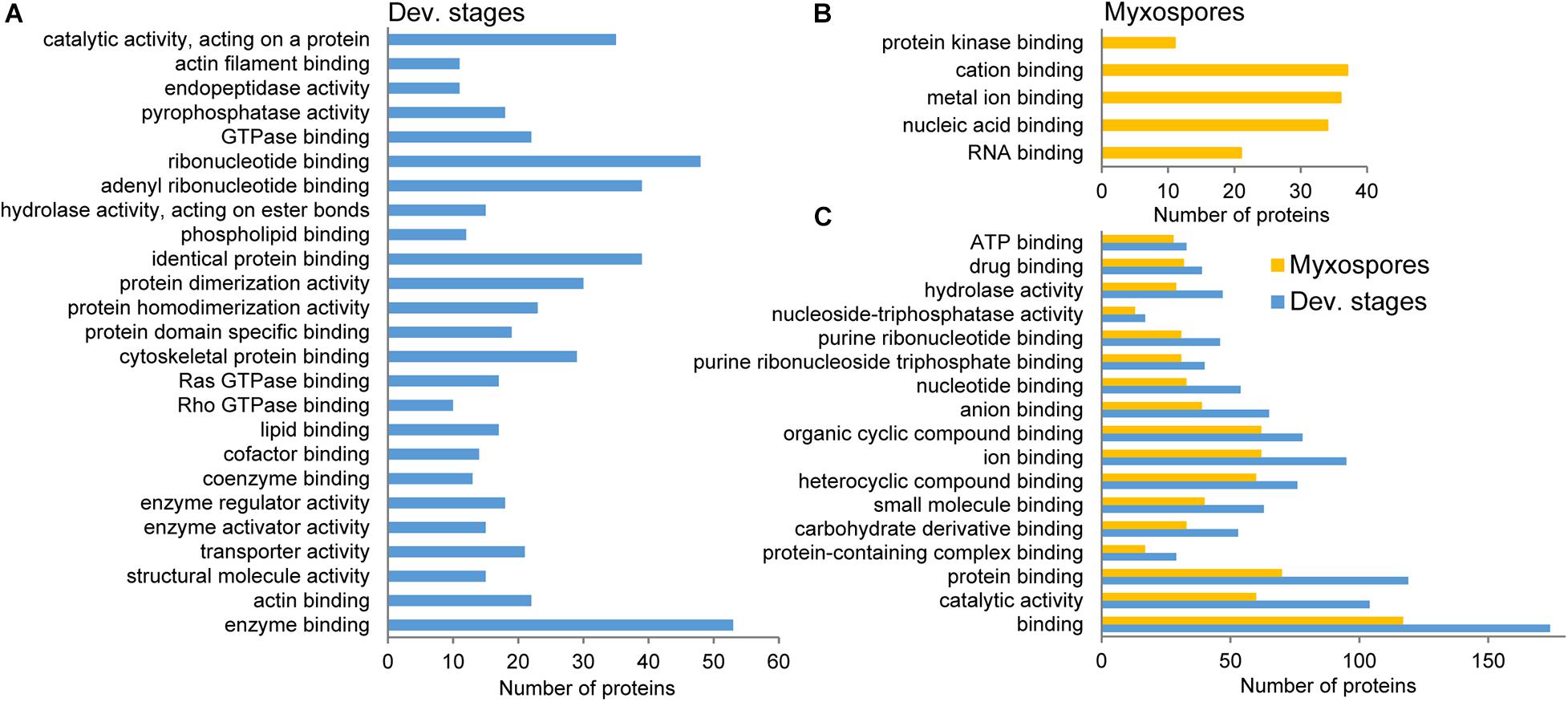
Figure 4. STRING GO enrichment analysis of molecular function of significantly expressed proteins in either developmental stages or mature myxospores. Only terms that were associated with at least 10 proteins are shown; FDR adjusted P < 0.05. (A) GO terms enriched in developmental stages. (B) GO terms enriched in myxospores. (C) Enriched GO terms shared by both sample types. Detailed results of the GO analysis are in Supplementary Table 4.
We next searched both proteomic datasets for all annotations and GO terms related to “actin” and “cytoskeleton” and used STRING to visualize the interaction (Figure 5A).
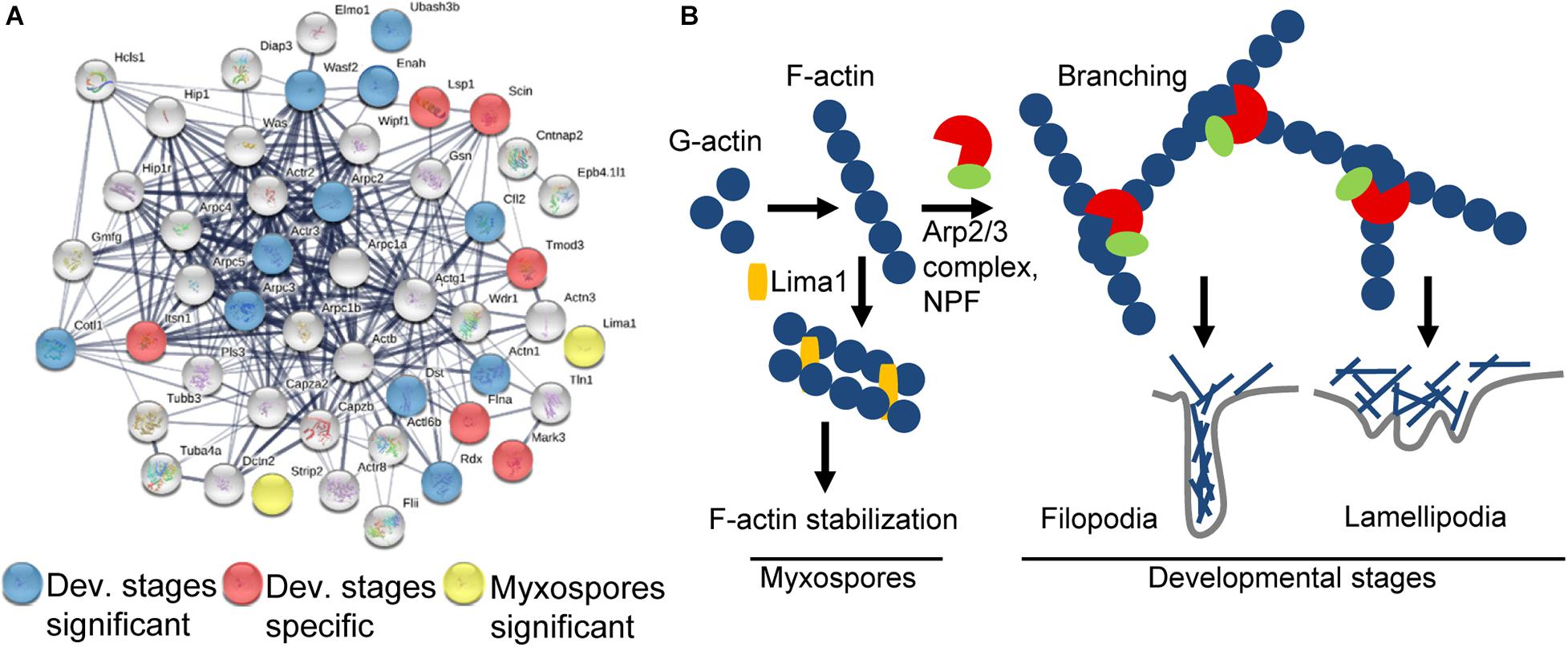
Figure 5. Schematic representation of the actin-related proteins. (A) STRING interaction map of proteins having “actin” or “cytoskeleton” GO term or annotation in both datasets. The STRING data were obtained using mouse protein homologs of C. shasta and the mouse symbols are near the nodes. Respectively, blue and red nodes represent proteins significantly expressed in or specific to developing spores, while yellow nodes represent proteins significantly expressed in mature myxospores. Gray nodes represent proteins common to both sample types. Strong associations are represented by thicker lines. (B) Key pathways found in mature myxospores and developmental stages. In developmental stages, F-actin is nucleated by Arp2/3 and NPFs to create branched actin, which promotes lamellipodia and filopodia formation that enable cell motility, whereas in mature myxospores F-actin is stabilized and nucleation is inhibited.
Findings From Developmental Stages
We identified 6 of the 7 subunits of the actin filament nucleator Arp2/3 complex, of which Arpc2 and Arpc3 were enriched (Pizarro-Cerdá et al., 2017). We also identified WASP, WDR1, Wipf1, MDia2, and a WAVE family member WAF2, the latter was significantly expressed. These proteins are all nucleation-promoting factors (NPFs), which activate the Arp2/3 complex and promote migration by actin polymerization, thus creating branched actin filament networks required for formation of lamellipodia or filopodia (Figure 5B; Campellone and Welch, 2010). Proteins expressed only in developmental stages include: intersectin 1 (ITSN1), which stimulates actin nucleation (Tsyba et al., 2011), Tropomodulin 3 (TMOD3), which contributes to the formation of the short actin protofilament that, in turn, defines the geometry of the cell membrane (Yamashiro et al., 2012), and talin 1 (TLN1), which plays a central role in cell adhesion through interaction with β integrin and F-actin (Goult et al., 2018). Among other significantly expressed proteins were ENAH, which promotes filopodia formation (Krugmann et al., 2001), actinin α1 (ACTN1), an F-actin cross-linking protein, coactosin-like protein (COTL1) that binds F-actin (Provost et al., 2001), adseverin (SCIN/ADSV), a Ca-dependent actin filament-severing protein affecting the organization of the microfilament network underneath the plasma membrane (Chumnarnsilpa et al., 2009), and Cofilin (CFL2), which controls actin polymerization and depolymerization at the leading edge of motile cells (DesMarais et al., 2005). Other significantly expressed proteins related to actin were the exocyst component Exo70, which favors membrane protrusion by promoting WAVE-Arp2/3 interaction (Zuo et al., 2006; Liu et al., 2012), 14-3-3ζ that, among its broad signaling functions, regulates cytoskeleton dynamics (Angrand et al., 2006), and two myosin proteins that target actin protrusions (Coluccio, 2020), MYO1E and MYO3B, the latter was specifically expressed only in developing spores. Additionally, under the enriched GO term “small GTPase proteins,” we found proteins related to cell migration, cell adhesion and cytoskeleton assembly, such as coronin 1C, serine/threonine-protein kinase N2 (Pkn2), TBC1 domain containing kinase (TBCK) and Rho GDP dissociation inhibitor α and β (ARHGDIA and B) that, through modulation of Rho proteins, play a role in cell motility regulation. In other enriched GO terms such as “lipid binding” we identified proteins that are related to membrane trafficking belonging to the annexin family and the sorting nexins group, and under the GO term “endopeptidase activity” we identified proteins such as cathepsin K, L and Z, and endopeptidases from the proteasome apparatus.
Findings From Mature Myxospores
Only two enriched actin-related proteins were identified: LIM domain and actin-binding 1 (LIMA1/EPLIN), a cytoskeleton-associated protein that inhibits actin filament depolymerization and cross-links filaments in bundles (Maul et al., 2003), and striatin-interacting phosphatase 2 (STRIP2), which plays a role in the regulation of cell shape and cytoskeletal organization (Bai et al., 2011). Additionally, under the enriched term “nucleic acid binding,” we found LIM domain binding 3 (Ldb3), which modulates protein-protein interaction domains through its PDZ domain, two proteins from the paired box family, OTX homeodomain protein, Piwil2, which binds RNA, five proteins (CNOT7, DDX6, MTREX/Skiv2l2, EXOS1, EXOS2) from the RNA degradation pathway and others.
Enriched GO terms that were shared between the developmental stages and mature myxospores were mostly related to fundamental DNA and RNA processes (Figure 4C and Supplementary Table 4).
Discussion
In the current study, we characterized and compared the proteomic profiles of different stages in the life cycle of C. shasta: developmental (proliferative, pre-sporogonic, and sporogonic stages) from within the fish host (Bartholomew et al., 1989a,b), and the resulting mature myxospores, which are viable outside the host to transmit infection. We used a proteomic approach because the mature spores are considered to be relatively transcriptionally silent and the proteome reflects an active state of all stages. Our findings demonstrate diverse proteins in developmental stages resulting from many metabolic processes, in comparison with the myxospores and provide the first reference map of the proteome of mature myxospores. We identified 2,296 protein groups, including proteins specifically expressed in each stage, and 357 protein groups with statistically significant stage-specific expression.
Our data show sporogenesis is characterized by actin polymerization and reorganization, which provide the driving force for the cell motility documented in developing parasites (Alama-Bermejo et al., 2020). The actin cytoskeleton plays a key role in most dynamic cellular processes, including cell shape changes, migration and division (Svitkina, 2018). In myxozoans, cell motility has been described during sporoplasm invasion into the host, on the route to the target tissue and during sporogenesis (Feist et al., 2015; Adriano and Okamura, 2017). Detailed ultrastructural analysis during Ceratomyxa puntazzi sporogenesis revealed an F-actin-rich cytoskeleton in filopodia that caused directional motility of the parasite (Alama-Bermejo et al., 2012), whereas in Sphaerospora molnari two types of actin were shown to be involved in cell motility during proliferation (Hartigan et al., 2016a). In highly virulent C. shasta genotype IIR, motility is due to high expression of RhoA, β-actin, and the adhesion-related genes integrin-β and talin (Alama-Bermejo et al., 2019). In the pre-sporogonic and sporogonic developmental stages, we identified the Arp2/3 complex of highly conserved nucleation proteins that, together with NPF proteins, generate branched actin networks. Additionally, we identified the homologous myxozoan molecular machinery that regulates and creates the actin filament network to form lamellipodia or filopodia. Our findings also support previous observation of the involvement of endopeptidases during myxozoan sporogenesis (Kelley et al., 2003; Alama-Bermejo et al., 2020).
In mature myxospores, we identified proteins homologous with those that enhance F-actin stabilization in mammals, including two LIM domain-containing proteins, LIMA1 and Ldb3, which are characterized by their interaction with F-actin (Kadrmas and Beckerle, 2004). Particularly interesting is LIMA1, which was first identified when it was downregulated in cancer cells (Maul and Chang, 1999) and later identified as a key protein that prevents actin depolymerization and functions during filament bundle assembly and stabilization (Maul et al., 2003; Abe and Takeichi, 2008; Taha et al., 2019). In addition, we identified STRIP2, which was shown in Drosophila to increase the level of cortical actin filaments (Bai et al., 2011). The high abundance of these proteins in the myxospore is consistent with the final rigid structure of the spores, demonstrating the transformation of the dynamic actin cytoskeleton network during sporogenesis to a stabilized F-actin structure. These findings also open the door for comparisons with actinospore stages in the annelid and with the sister lineage Malacosporea, which has less robust spore stages (Fiala et al., 2015).
In summary, we characterized the proteome of C. shasta during development in the fish host, and identified differences between motile proliferative stages and the mature myxospore, which is the robust transmission stage essential to continuing the myxozoan life cycle.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD022770 and 10.6019/PXD022770.
Ethics Statement
The animal study was reviewed and approved by Oregon State University’s Institutional Animal Care and Use Committee (IACUC; protocol #5040).
Author Contributions
GA-B, SA, JB, and TL conceived and designed the experiments. GA-B, SA, and VB performed the experiments. MO-L, AM, and TL analyzed the data. KM-L critically reviewed the manuscript. TL wrote the manuscript with input from all authors.
Funding
This research was supported by Research grant no. IS-5001-17C from BARD, The United States-Israel Binational Agricultural Research and Development Fund, by grants nos. 47496 and 43183 from United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, and by the Israel Science Foundation grant no. 43116.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Bioinformatics Facility at the University of Haifa and the Smoler Proteomics Center at the Technion for their assistance in the bioinformatics analysis and proteomic profiling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.632700/full#supplementary-material
Supplementary Table 1 | List of proteins identified in the study including annotation and proteomic data.
Supplementary Table 2 | Protein sequence of the identified proteins.
Supplementary Table 3 | List of proteins shown in Figure 3.
Supplementary Table 4 | GO analysis data in Figure 4.
Footnotes
References
Abe, K., and Takeichi, M. (2008). EPLIN mediates linkage of the cadherin–catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. U.S.A. 105, 13–19. doi: 10.1073/pnas.0710504105
Adriano, E., and Okamura, B. (2017). Motility, morphology and phylogeny of the plasmodial worm, Ceratomyxa vermiformis n. sp.(Cnidaria: Myxozoa: Myxosporea). Parasitology 144, 158–168.
Alama-Bermejo, G., Bron, J. E., Raga, J. A., and Holzer, A. S. (2012). 3D morphology, ultrastructure and development of Ceratomyxa puntazzi stages: first insights into the mechanisms of motility and budding in the Myxozoa. PLoS One 7:e32679. doi: 10.1371/journal.pone.0032679
Alama-Bermejo, G., Holzer, A. S., and Bartholomew, J. L. (2019). Myxozoan adhesion and virulence: Ceratonova shasta on the move. Microorganisms 7:397.
Alama-Bermejo, G., Meyer, E., Atkinson, S. D., Holzer, A. S., Wiśniewska, M. M., Kolísko, M., et al. (2020). Transcriptome-wide comparisons and virulence gene polymorphisms of host-associated genotypes of the cnidarian parasite Ceratonova shasta in salmonids. Genome Biol. Evol. 12, 1258–1276.
Angrand, P. O., Segura, I., Völkel, P., Ghidelli, S., Terry, R., Brajenovic, M., et al. (2006). Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol. Cell. Proteomics 5, 2211–2227. doi: 10.1074/mcp.M600147-MCP200
Atkinson, S. D., and Bartholomew, J. L. (2010). Disparate infection patterns of Ceratomyxa shasta (Myxozoa) in rainbow trout (Oncorhynchus mykiss) and Chinook salmon (Oncorhynchus tshawytscha) correlate with internal transcribed spacer-1 sequence variation in the parasite. Inter. J. Parasit. 40, 599–604. doi: 10.1016/j.ijpara.2009.10.010
Atkinson, S. D., Bartholomew, J. L., and Lotan, T. (2018a). Myxozoans: ancient metazoan parasites find a home in phylum Cnidaria. Zoology 129, 66–68. doi: 10.1016/j.zool.2018.06.005
Atkinson, S. D., Hallett, S. L., and Bartholomew, J. L. (2018b). Genotyping of individual Ceratonova shasta (Cnidaria: Myxosporea) myxospores reveals intra-spore ITS-1 variation and invalidates the distinction of genotypes II and III. Parasitology 145, 1588–1593.
Bai, S. W., Herrera-Abreu, M. T., Rohn, J. L., Racine, V., Tajadura, V., Suryavanshi, N., et al. (2011). Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC Biol. 9:54. doi: 10.1186/1741-7007-9-54
Bartholomew, J. L., Rohovec, J. S., and Fryer, J. L. (1989a). Development, characterization, and use of monoclonal and polyclonal antibodies against the myxosporean, Ceratomyxa shasta. J. Eukaryot. Microbiol. 36, 397–401. doi: 10.1111/j.1550-7408.1989.tb05534.x
Bartholomew, J. L., Smith, C., Rohovec, J., and Fryer, J. (1989b). Characterization of a host response to the myxosporean parasite, Ceratomyxa shasta (Noble), by histology, scanning electron microscopy and immunological techniques. J. Fish Dis. 12, 509–522.
Bartholomew, J. L., Whipple, M. J., Stevens, D. G., and Fryer, J. L. (1997). The life cycle of Ceratomyxa shasta, a myxosporean parasite of salmonids, requires a freshwater polychaete as an alternate host. J. Parasitol. 83, 859–868. doi: 10.2307/3284281
Ben-David, J., Atkinson, S. D., Pollak, Y., Yossifon, G., Shavit, U., Bartholomew, J. L., et al. (2016). Myxozoan polar tubules display structural and functional variation. Parasit. Vectors 9:549. doi: 10.1186/s13071-016-1819-4
Bjork, S. J., and Bartholomew, J. L. (2010). Invasion of Ceratomyxa shasta (Myxozoa) and comparison of migration to the intestine between susceptible and resistant fish hosts. Int. J. Parasitol. 40, 1087–1095. doi: 10.1016/j.ijpara.2010.03.005
Campellone, K. G., and Welch, M. D. (2010). A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251. doi: 10.1038/nrm2867
Canning, E. U., and Okamura, B. (2003). Biodiversity and evolution of the Myxozoa. Adv. Parasitol. 56, 43–131.
Chumnarnsilpa, S., Lee, W. L., Nag, S., Kannan, B., Larsson, M., Burtnick, L. D., et al. (2009). The crystal structure of the C-terminus of adseverin reveals the actin-binding interface. Proc. Natl. Acad. Sci. U.S.A. 106, 13719–13724. doi: 10.1073/pnas.0812383106
Coluccio, L. M. (2020). “Myosins and disease,” in Myosins: A Superfamily of Molecular Motors, ed. L. M. Coluccio (Cham: Springer International Publishing), 245–316.
DesMarais, V., Ghosh, M., Eddy, R., and Condeelis, J. (2005). Cofilin takes the lead. J. Cell Sci. 118, 19–26.
Feist, S. W., Morris, D. J., Alama-Bermejo, G., and Holzer, A. S. (2015). “Cellular processes in myxozoans,” in Myxozoan Evolution, Ecology and Development, eds B. Okamura, A. Gruhl, and J. Bartholomew (Cham: Springer), 139–154.
Fiala, I., Bartošová-Sojková, P., and Whipps, C. M. (2015). “Classification and phylogenetics of Myxozoa,” in Myxozoan Evolution, Ecology and Development, eds B. Okamura, A. Gruhl, and J. Bartholomew (Cham: Springer), 85–110.
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152.
Goult, B. T., Yan, J., and Schwartz, M. A. (2018). Talin as a mechanosensitive signaling hub. J. Cell Biol. 217, 3776–3784. doi: 10.1083/jcb.201808061
Guo, Q., Li, D., Zhai, Y., and Gu, Z. (2020). CCPRD: a novel analytical framework for the comprehensive proteomic reference database construction of NonModel organisms. ACS Omega 5, 15370–15384. doi: 10.1021/acsomega.0c01278
Hallett, S. L., and Bartholomew, J. L. (2011). “Myxobolus cerebralis and Ceratomyxa shasta,” in Fish Parasites: Pathobiology and Protection, eds P. T. K. Woo, and K. Buckmann (Wallingford: CAB International).
Hallett, S. L., Hartigan, A., and Atkinson, S. D. (2015). “Myxozoans on the move: dispersal modes, exotic species and emerging diseases,” in Myxozoan Evolution, Ecology and Development, eds B. Okamura, A. Gruhl, and J. L. Bartholomew (Cham: Springer), 343–362.
Hallett, S. L., Ray, R. A., Hurst, C. N., Holt, R. A., Buckles, G. R., Atkinson, S. D., et al. (2012). Density of the waterborne parasite Ceratomyxa shasta and its biological effects on salmon. Appl. Environ. Microbiol. 78, 3724–3731.
Hartigan, A., Estensoro, I., Vancová, M., Bílý, T., Patra, S., Eszterbauer, E., et al. (2016a). New cell motility model observed in parasitic cnidarian Sphaerospora molnari (Myxozoa:Myxosporea) blood stages in fish. Sci. Rep. 6:39093. doi: 10.1038/srep39093
Hartigan, A., Wilkinson, M., Gower, D. J., Streicher, J. W., Holzer, A. S., and Okamura, B. (2016b). Myxozoan infections of caecilians demonstrate broad host specificity and indicate a link with human activity. Int. J. Parasitol. 46, 375–381. doi: 10.1016/j.ijpara.2016.02.001
Hartikainen, H., Bass, D., Briscoe, A. G., Knipe, H., Green, A. J., and Okamura, B. (2016). Assessing myxozoan presence and diversity using environmental DNA. Int. J. Parasitol. 46, 781–792. doi: 10.1016/j.ijpara.2016.07.006
Jensen, L. J., Kuhn, M., Stark, M., Chaffron, S., Creevey, C., Muller, J., et al. (2009). STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37(Suppl. 1), D412–D416. doi: 10.1093/nar/gkn760
Kadrmas, J. L., and Beckerle, M. C. (2004). The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920–931. doi: 10.1038/nrm1499
Kelley, G. O., Adkison, M. A., Leutenegger, C. M., and Hedrick, R. P. (2003). Myxobolus cerebralis: identification of a cathepsin Z-like protease gene (MyxCP-1) expressed during parasite development in rainbow trout, Oncorhynchus mykiss. Exp. Parasitol. 105, 201–210. doi: 10.1016/j.exppara.2003.12.004
Krugmann, S., Jordens, I., Gevaert, K., Driessens, M., Vandekerckhove, J., and Hall, A. (2001). Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr. Biol. 11, 1645–1655. doi: 10.1016/S0960-9822(01)00506-1
Liu, J., Zhao, Y., Sun, Y., He, B., Yang, C., Svitkina, T., et al. (2012). Exo70 stimulates the Arp2/3 complex for lamellipodia formation and directional cell migration. Curr. Biol. 22, 1510–1515. doi: 10.1016/j.cub.2012.05.055
Lom, J., and Dyková, I. (2006). Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathogenic species. Folia Parasitol. 53, 1–36. doi: 10.14411/fp.2006.001
Mackenzie, K., and Kalavati, C. (2014). Myxosporean parasites of marine fishes: their distribution in the world’s oceans. Parasitology 141, 1709–1717. doi: 10.1017/S0031182014001425
Maul, R. S., and Chang, D. D. (1999). EPLIN, epithelial protein lost in neoplasm. Oncogene 18, 7838–7841. doi: 10.1038/sj.onc.1203206
Maul, R. S., Song, Y., Amann, K. J., Gerbin, S. C., Pollard, T. D., and Chang, D. D. (2003). EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J. Cell Biol. 160, 399–407. doi: 10.1083/jcb.200212057
Okamura, B., Gruhl, A., and Bartholomew, J. L. (2015). Myxozoan Evolution, Ecology and Development. Cham: Springer.
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Piriatinskiy, G., Atkinson, S. D., Park, S., Morgenstern, D., Brekhman, V., Yossifon, G., et al. (2017). Functional and proteomic analysis of Ceratonova shasta (Cnidaria: Myxozoa) polar capsules reveals adaptations to parasitism. Sci. Rep. 7:9010. doi: 10.1038/s41598-017-09955-y
Pizarro-Cerdá, J., Chorev, D. S., Geiger, B., and Cossart, P. (2017). The diverse family of Arp2/3 complexes. Trends Cell Biol. 27, 93–100. doi: 10.1016/j.tcb.2016.08.001
Provost, P., Doucet, J., Stock, A., Gerisch, G., Samuelsson, B., and Radmark, O. (2001). Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem. J. 359, 255–263. doi: 10.1042/0264-6021:3590255
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Sterud, E., Forseth, T., Ugedal, O., Poppe, T. T., Jørgensen, A., Bruheim, T., et al. (2007). Severe mortality in wild Atlantic salmon Salmo salar due to proliferative kidney disease (PKD) caused by Tetracapsuloides bryosalmonae (Myxozoa). Dis. Aquat. Organ. 77, 191–198. doi: 10.3354/dao01846
Stinson, M. E., Atkinson, S. D., and Bartholomew, J. L. (2018). Widespread distribution of Ceratonova shasta (Cnidaria: Myxosporea) genotypes indicates evolutionary adaptation to its salmonid fish hosts. J. Parasitol. 104, 645–650. doi: 10.1645/18-79
Svitkina, T. (2018). The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 10:a018267. doi: 10.1101/cshperspect.a018267
Taha, M., Aldirawi, M., März, S., Seebach, J., Odenthal-Schnittler, M., Bondareva, O., et al. (2019). EPLIN-α and -β Isoforms modulate endothelial cell dynamics through a spatiotemporally differentiated Interaction with actin. Cell Rep. 29, 1010–1026.e6. doi: 10.1016/j.celrep.2019.09.043
Tsyba, L., Nikolaienko, O., Dergai, O., Dergai, M., Novokhatska, O., Skrypkina, I., et al. (2011). Intersectin multidomain adaptor proteins: regulation of functional diversity. Gene 473, 67–75. doi: 10.1016/j.gene.2010.11.016
Wahli, T., Knuesel, R., Bernet, D., Segner, H., Pugovkin, D., Burkhardt-Holm, P., et al. (2002). Proliferative kidney disease in Switzerland: current state of knowledge. J. Fish Dis. 25, 491–500. doi: 10.1046/j.1365-2761.2002.00401.x
Wolf, K., and Markiw, M. E. (1984). Biology contravenes taxonomy in the Myxozoa: new discoveries show alternation of invertebrate and vertebrate hosts. Science 225, 1449–1452. doi: 10.1126/science.225.4669.1449
Yamashiro, S., Gokhin, D. S., Kimura, S., Nowak, R. B., and Fowler, V. M. (2012). Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton 69, 337–370. doi: 10.1002/cm.21031
Keywords: Cnidaria, Myxozoa, parasite, sporogenesis, cytoskeleton, actin
Citation: Brekhman V, Ofek-Lalzar M, Atkinson SD, Alama-Bermejo G, Maor-Landaw K, Malik A, Bartholomew JL and Lotan T (2021) Proteomic Analysis of the Parasitic Cnidarian Ceratonova shasta (Cnidaria: Myxozoa) Reveals Diverse Roles of Actin in Motility and Spore Formation. Front. Mar. Sci. 8:632700. doi: 10.3389/fmars.2021.632700
Received: 26 January 2021; Accepted: 26 March 2021;
Published: 15 April 2021.
Edited by:
Jorge Galindo-Villegas, Nord University, NorwayReviewed by:
Gu Zemao, Huazhong Agricultural University, ChinaJy Zhang, Qingdao Agricultural University, China
Copyright © 2021 Brekhman, Ofek-Lalzar, Atkinson, Alama-Bermejo, Maor-Landaw, Malik, Bartholomew and Lotan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tamar Lotan, lotant@univ.haifa.ac.il
 Vera Brekhman
Vera Brekhman Maya Ofek-Lalzar
Maya Ofek-Lalzar Stephen D. Atkinson
Stephen D. Atkinson Gema Alama-Bermejo
Gema Alama-Bermejo Keren Maor-Landaw
Keren Maor-Landaw Assaf Malik
Assaf Malik Jerri L. Bartholomew3
Jerri L. Bartholomew3  Tamar Lotan
Tamar Lotan